Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

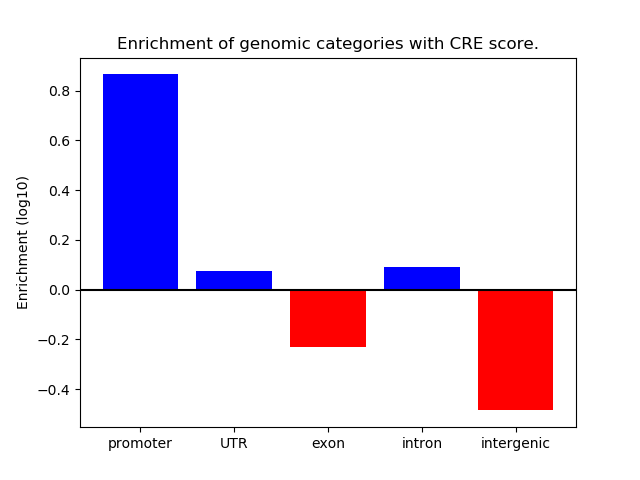
CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
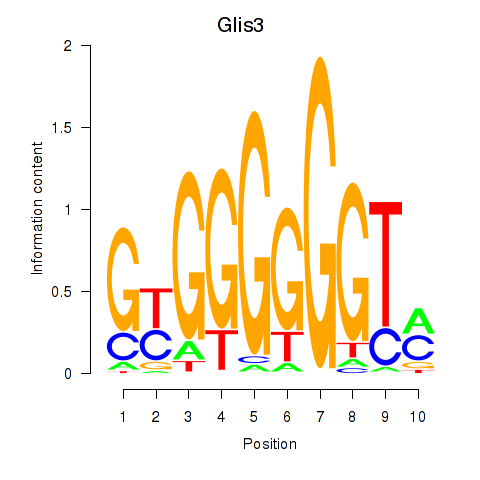
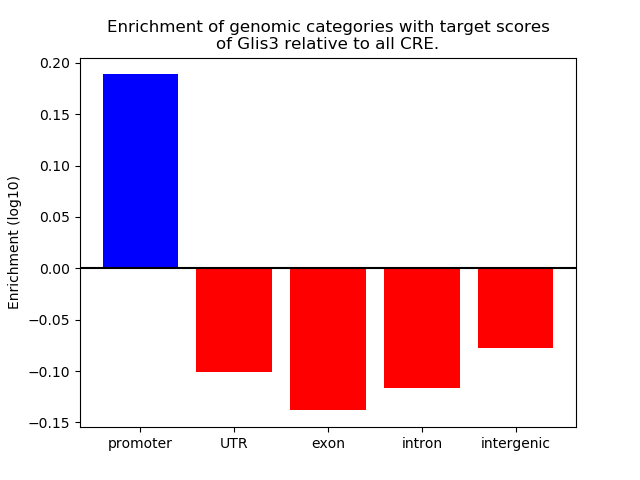
Results for Glis3
Z-value: 0.69
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.7 | Glis3 |
|
Glis3
|
ENSMUSG00000091294.1 | Glis3 |
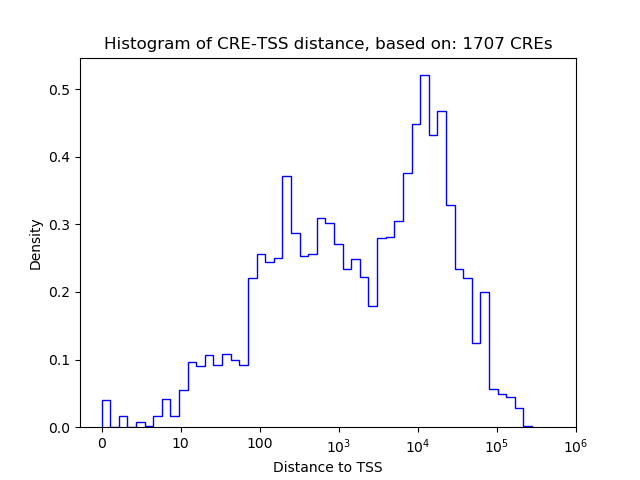
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Glis3 | chr19_28328619_28328770 | 2688 | 0.402656 | 0.57 | 6.3e-06 | Click! |
| Glis3 | chr19_28679781_28680338 | 18 | 0.971293 | 0.46 | 4.7e-04 | Click! |
| Glis3 | chr19_28641105_28641256 | 25505 | 0.170254 | 0.27 | 4.3e-02 | Click! |
| Glis3 | chr19_28380638_28380789 | 54707 | 0.160298 | 0.26 | 5.5e-02 | Click! |
| Glis3 | chr19_28436583_28436752 | 103753 | 0.071479 | 0.24 | 7.2e-02 | Click! |
Activity of the Glis3 motif across conditions
Conditions sorted by the z-value of the Glis3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
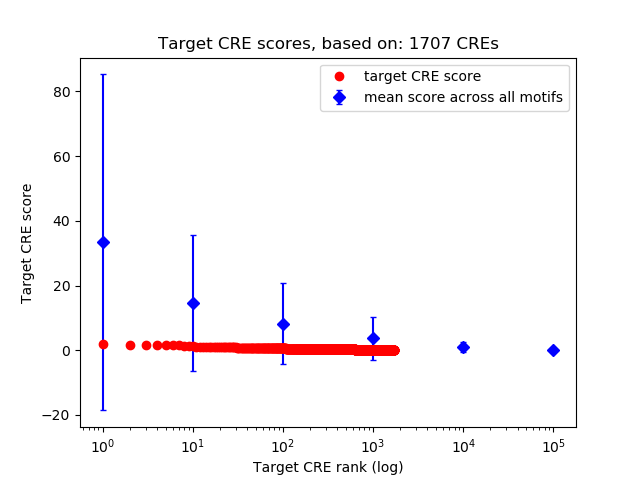
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_67471274_67471636 | 1.99 |
Gm33926 |
predicted gene, 33926 |
27522 |
0.17 |
| chr5_138997857_138998317 | 1.69 |
Pdgfa |
platelet derived growth factor, alpha |
717 |
0.64 |
| chr5_120005810_120006020 | 1.66 |
Gm38554 |
predicted gene, 38554 |
46672 |
0.13 |
| chr5_137348677_137348944 | 1.63 |
Ephb4 |
Eph receptor B4 |
1299 |
0.25 |
| chr12_3834339_3834973 | 1.62 |
Dnmt3a |
DNA methyltransferase 3A |
14979 |
0.17 |
| chr14_54971406_54971557 | 1.48 |
Gm29015 |
predicted gene 29015 |
1548 |
0.13 |
| chr3_69588417_69588708 | 1.47 |
B3galnt1 |
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
10398 |
0.17 |
| chr5_119836588_119836964 | 1.35 |
Tbx5 |
T-box 5 |
621 |
0.7 |
| chr15_93825328_93825719 | 1.23 |
Gm49445 |
predicted gene, 49445 |
485 |
0.86 |
| chr4_147969190_147969341 | 1.15 |
Nppb |
natriuretic peptide type B |
16523 |
0.09 |
| chr10_45181043_45181210 | 1.14 |
Popdc3 |
popeye domain containing 3 |
3028 |
0.27 |
| chr11_72041836_72042510 | 1.14 |
Pimreg |
PICALM interacting mitotic regulator |
141 |
0.94 |
| chr1_171189506_171189734 | 1.13 |
Pcp4l1 |
Purkinje cell protein 4-like 1 |
6648 |
0.09 |
| chr5_122100700_122101295 | 1.12 |
Myl2 |
myosin, light polypeptide 2, regulatory, cardiac, slow |
14 |
0.97 |
| chr14_25365371_25365579 | 1.12 |
Gm26660 |
predicted gene, 26660 |
13160 |
0.21 |
| chr16_44687184_44688343 | 1.10 |
Nepro |
nucleolus and neural progenitor protein |
36538 |
0.14 |
| chr2_30604975_30605527 | 1.07 |
Cstad |
CSA-conditional, T cell activation-dependent protein |
34 |
0.97 |
| chr2_32317120_32318698 | 1.05 |
Gm23363 |
predicted gene, 23363 |
356 |
0.45 |
| chr3_89391710_89392572 | 1.04 |
Zbtb7b |
zinc finger and BTB domain containing 7B |
72 |
0.62 |
| chr18_75378512_75379142 | 1.01 |
Smad7 |
SMAD family member 7 |
3913 |
0.25 |
| chr2_120094110_120094375 | 1.00 |
Sptbn5 |
spectrin beta, non-erythrocytic 5 |
8564 |
0.15 |
| chr17_29451348_29451678 | 1.00 |
Gm36199 |
predicted gene, 36199 |
2161 |
0.2 |
| chr14_62259091_62259242 | 0.99 |
Rnaseh2b |
ribonuclease H2, subunit B |
33423 |
0.15 |
| chr13_55510408_55510601 | 0.98 |
Pdlim7 |
PDZ and LIM domain 7 |
1484 |
0.19 |
| chr7_105580903_105581664 | 0.97 |
Apbb1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 |
5 |
0.95 |
| chr11_61484326_61485106 | 0.91 |
Mfap4 |
microfibrillar-associated protein 4 |
715 |
0.53 |
| chr2_33966954_33967150 | 0.90 |
Gm13404 |
predicted gene 13404 |
11194 |
0.21 |
| chr6_85254101_85254349 | 0.89 |
Sfxn5 |
sideroflexin 5 |
10708 |
0.16 |
| chr11_70468019_70468408 | 0.88 |
Zmynd15 |
zinc finger, MYND-type containing 15 |
7648 |
0.07 |
| chr5_125264626_125264993 | 0.86 |
Gm32585 |
predicted gene, 32585 |
8006 |
0.18 |
| chr8_102786608_102786857 | 0.82 |
Cdh11 |
cadherin 11 |
1090 |
0.52 |
| chr9_77352516_77352683 | 0.82 |
Mlip |
muscular LMNA-interacting protein |
370 |
0.84 |
| chr10_99534471_99534622 | 0.82 |
Gm48085 |
predicted gene, 48085 |
3523 |
0.24 |
| chr6_88937928_88938520 | 0.82 |
4933427D06Rik |
RIKEN cDNA 4933427D06 gene |
12459 |
0.13 |
| chr3_108085301_108086522 | 0.82 |
Gm12500 |
predicted gene 12500 |
65 |
0.8 |
| chr14_27114666_27114961 | 0.80 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
86 |
0.98 |
| chr14_21988166_21988548 | 0.78 |
Zfp503 |
zinc finger protein 503 |
1244 |
0.37 |
| chr11_80809988_80810207 | 0.77 |
Tmem98 |
transmembrane protein 98 |
78 |
0.97 |
| chr17_4866022_4866173 | 0.76 |
4930579D07Rik |
RIKEN cDNA 4930579D07 gene |
11615 |
0.2 |
| chr10_111594710_111594919 | 0.76 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
541 |
0.73 |
| chr7_79149006_79149317 | 0.76 |
Mfge8 |
milk fat globule-EGF factor 8 protein |
101 |
0.97 |
| chr8_14911749_14912202 | 0.74 |
Arhgef10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
237 |
0.92 |
| chr14_52014304_52014473 | 0.74 |
Zfp219 |
zinc finger protein 219 |
194 |
0.83 |
| chr7_142657124_142657560 | 0.74 |
Igf2 |
insulin-like growth factor 2 |
124 |
0.92 |
| chr7_126582804_126583424 | 0.74 |
Cln3 |
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
0 |
0.93 |
| chr2_35684118_35684369 | 0.73 |
Dab2ip |
disabled 2 interacting protein |
7751 |
0.24 |
| chr13_30782030_30782219 | 0.73 |
Gm29675 |
predicted gene, 29675 |
13102 |
0.19 |
| chr7_29453052_29453769 | 0.73 |
Sipa1l3 |
signal-induced proliferation-associated 1 like 3 |
52042 |
0.1 |
| chr16_44632721_44633371 | 0.72 |
Boc |
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
74149 |
0.08 |
| chr9_14596490_14596733 | 0.72 |
Amotl1 |
angiomotin-like 1 |
17975 |
0.12 |
| chr8_11357901_11358379 | 0.72 |
Col4a1 |
collagen, type IV, alpha 1 |
45314 |
0.1 |
| chr3_79690130_79690466 | 0.72 |
Gm26984 |
predicted gene, 26984 |
13422 |
0.15 |
| chr16_43503682_43504014 | 0.71 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
151 |
0.97 |
| chr1_39288126_39288277 | 0.70 |
Gm20428 |
predicted gene 20428 |
11636 |
0.17 |
| chr17_67170823_67170977 | 0.70 |
Ptprm |
protein tyrosine phosphatase, receptor type, M |
122915 |
0.05 |
| chr13_37629070_37629705 | 0.70 |
Gm40915 |
predicted gene, 40915 |
1784 |
0.24 |
| chr2_36104921_36105072 | 0.69 |
Lhx6 |
LIM homeobox protein 6 |
105 |
0.95 |
| chr2_28446566_28447382 | 0.68 |
Ppp1r26 |
protein phosphatase 1, regulatory subunit 26 |
108 |
0.95 |
| chr13_60737684_60738259 | 0.68 |
Dapk1 |
death associated protein kinase 1 |
16025 |
0.15 |
| chr6_53218861_53219048 | 0.67 |
Gm4872 |
predicted gene 4872 |
2210 |
0.36 |
| chr3_95687508_95688286 | 0.67 |
Adamtsl4 |
ADAMTS-like 4 |
20 |
0.96 |
| chr11_71747926_71748095 | 0.67 |
Wscd1 |
WSC domain containing 1 |
1910 |
0.3 |
| chr19_47885000_47885151 | 0.67 |
Gsto2 |
glutathione S-transferase omega 2 |
14813 |
0.13 |
| chr14_27622251_27622923 | 0.66 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
77 |
0.98 |
| chr12_45022163_45022314 | 0.66 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
9274 |
0.23 |
| chr10_81559146_81561402 | 0.65 |
Tle5 |
TLE family member 5, transcriptional modulator |
770 |
0.38 |
| chr4_66894714_66894865 | 0.65 |
Tlr4 |
toll-like receptor 4 |
66587 |
0.14 |
| chr14_120637683_120637943 | 0.65 |
Gm49008 |
predicted gene, 49008 |
4851 |
0.28 |
| chr7_139467729_139467880 | 0.65 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
13750 |
0.24 |
| chr12_16553856_16554148 | 0.64 |
Lpin1 |
lipin 1 |
8036 |
0.26 |
| chr5_122102023_122102402 | 0.64 |
Myl2 |
myosin, light polypeptide 2, regulatory, cardiac, slow |
415 |
0.79 |
| chr12_108371321_108371691 | 0.64 |
Eml1 |
echinoderm microtubule associated protein like 1 |
549 |
0.75 |
| chr17_44300705_44301078 | 0.64 |
Clic5 |
chloride intracellular channel 5 |
63785 |
0.12 |
| chr18_55815948_55816458 | 0.64 |
Gm8614 |
predicted gene 8614 |
22746 |
0.19 |
| chr5_122950982_122952145 | 0.63 |
Kdm2b |
lysine (K)-specific demethylase 2B |
427 |
0.77 |
| chr15_82961007_82961398 | 0.63 |
Mir3080 |
microRNA 3080 |
1906 |
0.23 |
| chr17_71039919_71040277 | 0.62 |
Myom1 |
myomesin 1 |
20507 |
0.12 |
| chr10_86371635_86371796 | 0.62 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
68861 |
0.08 |
| chr11_94883072_94883246 | 0.61 |
A430060F13Rik |
RIKEN cDNA A430060F13 gene |
21400 |
0.11 |
| chr1_74284190_74285456 | 0.60 |
Aamp |
angio-associated migratory protein |
85 |
0.51 |
| chr13_48260396_48261208 | 0.60 |
Id4 |
inhibitor of DNA binding 4 |
426 |
0.7 |
| chr3_89315223_89316179 | 0.60 |
Gm15998 |
predicted gene 15998 |
262 |
0.77 |
| chr17_26424377_26424632 | 0.60 |
Neurl1b |
neuralized E3 ubiquitin protein ligase 1B |
9539 |
0.14 |
| chr14_120705317_120705468 | 0.60 |
Rpl19-ps5 |
ribosomal protein L19, pseudogene 5 |
7686 |
0.21 |
| chr1_74025160_74025359 | 0.60 |
Tns1 |
tensin 1 |
8837 |
0.23 |
| chr15_97055229_97055380 | 0.59 |
Slc38a4 |
solute carrier family 38, member 4 |
652 |
0.81 |
| chrX_160418379_160418554 | 0.58 |
Adgrg2 |
adhesion G protein-coupled receptor G2 |
8826 |
0.22 |
| chr3_87971441_87971954 | 0.58 |
Nes |
nestin |
568 |
0.56 |
| chr13_63573352_63573844 | 0.58 |
Ptch1 |
patched 1 |
0 |
0.96 |
| chr14_54994259_54994475 | 0.58 |
Myh7 |
myosin, heavy polypeptide 7, cardiac muscle, beta |
176 |
0.85 |
| chr2_114052677_114053403 | 0.57 |
Actc1 |
actin, alpha, cardiac muscle 1 |
153 |
0.93 |
| chr1_90897082_90897304 | 0.57 |
Mlph |
melanophilin |
17892 |
0.14 |
| chr8_115250365_115250516 | 0.57 |
Gm22556 |
predicted gene, 22556 |
197527 |
0.03 |
| chrX_150814229_150814429 | 0.57 |
Maged2 |
melanoma antigen, family D, 2 |
16 |
0.97 |
| chr18_76469688_76469918 | 0.56 |
Gm50360 |
predicted gene, 50360 |
147979 |
0.04 |
| chr10_42614130_42614512 | 0.56 |
Ostm1 |
osteopetrosis associated transmembrane protein 1 |
30483 |
0.16 |
| chr13_41345306_41345621 | 0.56 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
13784 |
0.14 |
| chr6_126646041_126646221 | 0.56 |
Kcna1 |
potassium voltage-gated channel, shaker-related subfamily, member 1 |
253 |
0.93 |
| chr18_36017856_36019112 | 0.56 |
Nrg2 |
neuregulin 2 |
13457 |
0.16 |
| chr16_85803746_85803905 | 0.56 |
Adamts1 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
712 |
0.73 |
| chr3_96181091_96181472 | 0.55 |
Sv2a |
synaptic vesicle glycoprotein 2 a |
130 |
0.86 |
| chr19_36348119_36348437 | 0.54 |
Gm47735 |
predicted gene, 47735 |
20 |
0.5 |
| chr9_107593161_107593336 | 0.54 |
Ifrd2 |
interferon-related developmental regulator 2 |
621 |
0.32 |
| chr17_26865252_26865474 | 0.54 |
Gm20468 |
predicted gene 20468 |
3942 |
0.12 |
| chr11_61485173_61485597 | 0.54 |
Mfap4 |
microfibrillar-associated protein 4 |
46 |
0.96 |
| chr5_105346543_105346806 | 0.54 |
Gbp11 |
guanylate binding protein 11 |
202 |
0.94 |
| chr4_100101880_100102395 | 0.53 |
Ror1 |
receptor tyrosine kinase-like orphan receptor 1 |
6346 |
0.22 |
| chr14_31205580_31207467 | 0.53 |
Nisch |
nischarin |
177 |
0.89 |
| chr3_79907276_79907427 | 0.53 |
Gm36569 |
predicted gene, 36569 |
20545 |
0.17 |
| chr2_157058410_157059040 | 0.53 |
Soga1 |
suppressor of glucose, autophagy associated 1 |
18486 |
0.12 |
| chr11_80316377_80316533 | 0.53 |
Rhbdl3 |
rhomboid like 3 |
3235 |
0.24 |
| chr16_35215095_35215432 | 0.52 |
Gm25179 |
predicted gene, 25179 |
19660 |
0.19 |
| chr1_153172459_153172671 | 0.52 |
Lamc2 |
laminin, gamma 2 |
13482 |
0.2 |
| chr11_101232085_101232549 | 0.52 |
Ezh1 |
enhancer of zeste 1 polycomb repressive complex 2 subunit |
5854 |
0.08 |
| chr1_193734040_193734306 | 0.51 |
Gm21362 |
predicted gene, 21362 |
132836 |
0.05 |
| chr5_114671329_114671480 | 0.51 |
Trpv4 |
transient receptor potential cation channel, subfamily V, member 4 |
12983 |
0.12 |
| chr4_152273539_152274471 | 0.51 |
Gpr153 |
G protein-coupled receptor 153 |
227 |
0.89 |
| chr10_126927498_126927649 | 0.51 |
Atp23 |
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
26218 |
0.09 |
| chr3_73056881_73057482 | 0.51 |
Slitrk3 |
SLIT and NTRK-like family, member 3 |
238 |
0.92 |
| chr6_97487685_97488107 | 0.51 |
Frmd4b |
FERM domain containing 4B |
91 |
0.97 |
| chr11_97659525_97659953 | 0.51 |
Gm24158 |
predicted gene, 24158 |
588 |
0.37 |
| chr5_65106807_65107827 | 0.50 |
Klhl5 |
kelch-like 5 |
222 |
0.92 |
| chr18_82912875_82914544 | 0.50 |
Zfp516 |
zinc finger protein 516 |
923 |
0.51 |
| chr1_134333064_134333269 | 0.50 |
Ppfia4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
238 |
0.89 |
| chr10_63457395_63457598 | 0.49 |
Ctnna3 |
catenin (cadherin associated protein), alpha 3 |
14 |
0.98 |
| chr19_55757537_55757688 | 0.49 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
14767 |
0.27 |
| chr9_102689518_102690683 | 0.49 |
4930533D04Rik |
RIKEN cDNA 4930533D04 gene |
2785 |
0.18 |
| chr6_128028583_128028766 | 0.49 |
Tspan9 |
tetraspanin 9 |
5918 |
0.2 |
| chr13_53237536_53237687 | 0.49 |
Ror2 |
receptor tyrosine kinase-like orphan receptor 2 |
38047 |
0.19 |
| chr10_83648138_83649089 | 0.49 |
Appl2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
10 |
0.98 |
| chr9_50722434_50722597 | 0.48 |
Dixdc1 |
DIX domain containing 1 |
5423 |
0.13 |
| chr1_156779038_156779543 | 0.48 |
Gm15428 |
predicted pseudogene 15428 |
14222 |
0.15 |
| chr19_4711655_4711852 | 0.48 |
Sptbn2 |
spectrin beta, non-erythrocytic 2 |
586 |
0.61 |
| chr8_47413184_47413712 | 0.48 |
Stox2 |
storkhead box 2 |
389 |
0.87 |
| chr8_71498904_71499355 | 0.48 |
Gtpbp3 |
GTP binding protein 3 |
9670 |
0.07 |
| chr8_88567927_88568131 | 0.47 |
Gm45496 |
predicted gene 45496 |
7996 |
0.2 |
| chr5_138912148_138912327 | 0.47 |
Pdgfa |
platelet derived growth factor, alpha |
82044 |
0.07 |
| chr11_59009654_59009805 | 0.47 |
Gm10435 |
predicted gene 10435 |
6819 |
0.09 |
| chr8_11331273_11331424 | 0.47 |
Col4a1 |
collagen, type IV, alpha 1 |
18522 |
0.15 |
| chr11_20858852_20859410 | 0.47 |
Gm22807 |
predicted gene, 22807 |
11250 |
0.15 |
| chr6_83794439_83795940 | 0.47 |
Nagk |
N-acetylglucosamine kinase |
19 |
0.94 |
| chr12_112147478_112147709 | 0.47 |
Kif26a |
kinesin family member 26A |
1385 |
0.33 |
| chr2_152048577_152049360 | 0.47 |
AA387200 |
expressed sequence AA387200 |
27840 |
0.11 |
| chr15_74487345_74487780 | 0.46 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
28633 |
0.16 |
| chr5_77358327_77358734 | 0.46 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
18162 |
0.13 |
| chr5_119794079_119794230 | 0.46 |
1700021F13Rik |
RIKEN cDNA 1700021F13 gene |
13513 |
0.17 |
| chr16_32854480_32854631 | 0.46 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
13783 |
0.13 |
| chr8_57321535_57322223 | 0.46 |
Hand2os1 |
Hand2, opposite strand 1 |
651 |
0.47 |
| chr4_117124515_117125246 | 0.46 |
Btbd19 |
BTB (POZ) domain containing 19 |
286 |
0.73 |
| chr13_56703202_56704180 | 0.46 |
Smad5 |
SMAD family member 5 |
23 |
0.98 |
| chr18_42978690_42978841 | 0.45 |
Ppp2r2b |
protein phosphatase 2, regulatory subunit B, beta |
40533 |
0.15 |
| chr11_98935983_98936905 | 0.45 |
Rara |
retinoic acid receptor, alpha |
1254 |
0.3 |
| chr9_56505269_56505564 | 0.45 |
Gm47175 |
predicted gene, 47175 |
1245 |
0.42 |
| chr10_58233022_58233496 | 0.45 |
Duxf3 |
double homeobox family member 3 |
584 |
0.62 |
| chr1_58030580_58030815 | 0.45 |
Aox1 |
aldehyde oxidase 1 |
733 |
0.64 |
| chr18_35918777_35919433 | 0.45 |
Gm50405 |
predicted gene, 50405 |
4190 |
0.13 |
| chr3_94582467_94582919 | 0.45 |
Snx27 |
sorting nexin family member 27 |
14 |
0.95 |
| chr1_156780969_156781575 | 0.45 |
Gm15428 |
predicted pseudogene 15428 |
12240 |
0.15 |
| chr10_38986094_38986316 | 0.45 |
Lama4 |
laminin, alpha 4 |
20533 |
0.22 |
| chr11_7055456_7055607 | 0.45 |
Adcy1 |
adenylate cyclase 1 |
7958 |
0.26 |
| chr10_62016778_62017263 | 0.44 |
Gm47919 |
predicted gene, 47919 |
6384 |
0.19 |
| chr9_118694402_118694558 | 0.44 |
Itga9 |
integrin alpha 9 |
8645 |
0.24 |
| chr13_18043214_18043365 | 0.44 |
Gm6575 |
predicted gene 6575 |
4728 |
0.14 |
| chr3_34642143_34642294 | 0.44 |
Gm42692 |
predicted gene 42692 |
1046 |
0.36 |
| chr2_166196150_166196314 | 0.44 |
Gm11466 |
predicted gene 11466 |
5370 |
0.17 |
| chr2_65567574_65567908 | 0.44 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
114 |
0.97 |
| chr7_67910990_67911267 | 0.44 |
Gm44666 |
predicted gene 44666 |
24543 |
0.16 |
| chr17_4173988_4174168 | 0.44 |
4930548J01Rik |
RIKEN cDNA 4930548J01 gene |
51965 |
0.17 |
| chr11_118360927_118361104 | 0.44 |
Timp2 |
tissue inhibitor of metalloproteinase 2 |
5275 |
0.16 |
| chr5_124964039_124964493 | 0.44 |
Rflna |
refilin A |
38955 |
0.13 |
| chr18_80712591_80713347 | 0.44 |
Nfatc1 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
89 |
0.97 |
| chr1_179667564_179668717 | 0.44 |
Sccpdh |
saccharopine dehydrogenase (putative) |
70 |
0.98 |
| chr19_16132209_16133500 | 0.43 |
Gnaq |
guanine nucleotide binding protein, alpha q polypeptide |
23 |
0.98 |
| chr9_8003315_8004975 | 0.43 |
Yap1 |
yes-associated protein 1 |
245 |
0.79 |
| chr5_132334819_132335092 | 0.43 |
Gm42989 |
predicted gene 42989 |
37726 |
0.16 |
| chr10_80936253_80937073 | 0.43 |
Gadd45b |
growth arrest and DNA-damage-inducible 45 beta |
5851 |
0.1 |
| chr13_80962745_80963559 | 0.43 |
9330111N05Rik |
RIKEN cDNA 9330111N05 gene |
264 |
0.71 |
| chr15_83487585_83487761 | 0.43 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
23067 |
0.11 |
| chr5_65107840_65108141 | 0.43 |
Klhl5 |
kelch-like 5 |
451 |
0.8 |
| chr3_89280469_89281651 | 0.43 |
Efna1 |
ephrin A1 |
82 |
0.91 |
| chr8_82967937_82968088 | 0.43 |
Gm18212 |
predicted gene, 18212 |
15500 |
0.24 |
| chr1_133255964_133256140 | 0.43 |
Gm19461 |
predicted gene, 19461 |
300 |
0.64 |
| chr16_33580751_33581254 | 0.43 |
Slc12a8 |
solute carrier family 12 (potassium/chloride transporters), member 8 |
15560 |
0.25 |
| chr2_59824280_59824521 | 0.43 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
17340 |
0.21 |
| chr15_80000729_80000880 | 0.42 |
Mir7213 |
microRNA 7213 |
1778 |
0.23 |
| chr7_74012764_74012935 | 0.42 |
St8sia2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
825 |
0.64 |
| chr15_99771905_99772913 | 0.42 |
Cers5 |
ceramide synthase 5 |
53 |
0.94 |
| chr10_97565444_97565849 | 0.42 |
Lum |
lumican |
518 |
0.78 |
| chr4_120131881_120132032 | 0.42 |
Edn2 |
endothelin 2 |
29250 |
0.18 |
| chr10_56376223_56376507 | 0.42 |
Gja1 |
gap junction protein, alpha 1 |
965 |
0.58 |
| chr10_79926423_79927890 | 0.42 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
17 |
0.92 |
| chr15_101294156_101294470 | 0.42 |
Smim41 |
small integral membrane protein 41 |
1081 |
0.33 |
| chr13_30659801_30660671 | 0.42 |
Dusp22 |
dual specificity phosphatase 22 |
135 |
0.97 |
| chr18_60656319_60656539 | 0.42 |
Synpo |
synaptopodin |
3713 |
0.22 |
| chr4_84199256_84199407 | 0.42 |
Gm12414 |
predicted gene 12414 |
69042 |
0.11 |
| chr6_5802451_5802946 | 0.42 |
Dync1i1 |
dynein cytoplasmic 1 intermediate chain 1 |
45279 |
0.2 |
| chr16_26185747_26186199 | 0.42 |
P3h2 |
prolyl 3-hydroxylase 2 |
80189 |
0.11 |
| chr12_29532917_29533068 | 0.41 |
Myt1l |
myelin transcription factor 1-like |
393 |
0.85 |
| chr8_40975906_40976229 | 0.41 |
Mtus1 |
mitochondrial tumor suppressor 1 |
24138 |
0.14 |
| chr8_70493079_70493663 | 0.41 |
Crlf1 |
cytokine receptor-like factor 1 |
10 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.4 | 1.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.3 | 1.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.6 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.9 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.9 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.2 | 0.7 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.2 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 0.5 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.6 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.5 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.1 | 0.4 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.3 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.7 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.3 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.7 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.2 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.1 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 | 0.2 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.6 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 0.2 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.3 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.3 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.3 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.2 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.1 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) |
| 0.1 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.3 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:1901844 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:1903365 | regulation of fear response(GO:1903365) positive regulation of fear response(GO:1903367) regulation of behavioral fear response(GO:2000822) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0061196 | fungiform papilla development(GO:0061196) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 1.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0043328 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.0 | GO:0098815 | modulation of excitatory postsynaptic potential(GO:0098815) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.6 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0061377 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.2 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.3 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.0 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:0045762 | positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:1904377 | positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 1.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.4 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.5 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.6 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.5 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.3 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0018591 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0004394 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |