Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
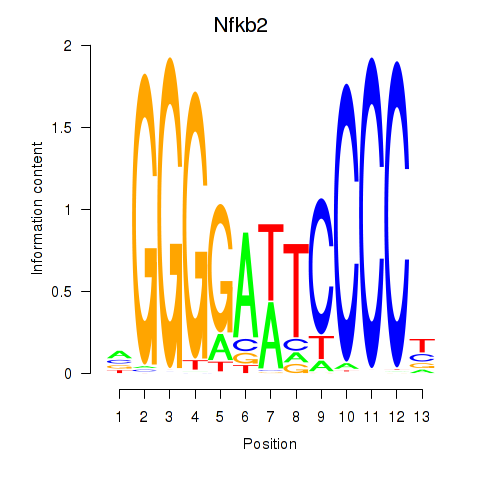
Results for Nfkb2
Z-value: 1.14
Transcription factors associated with Nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfkb2
|
ENSMUSG00000025225.8 | Nfkb2 |
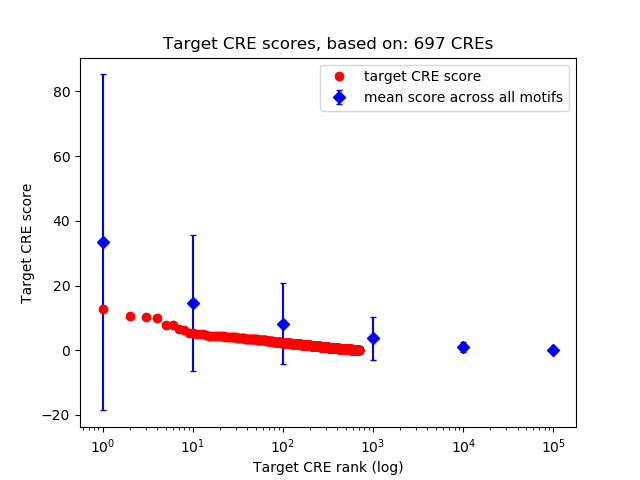
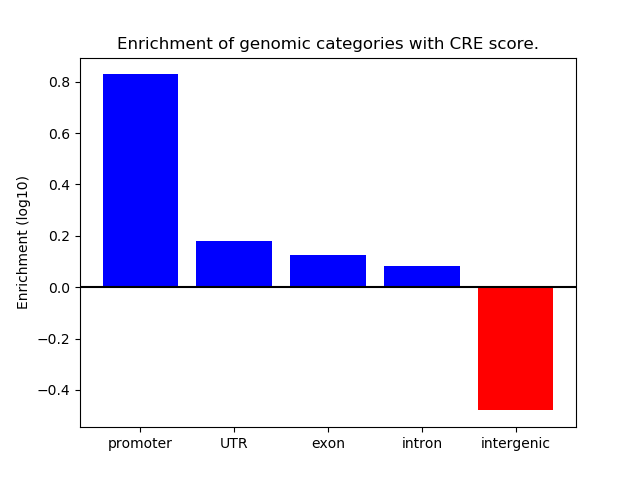
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Nfkb2 | chr19_46305520_46305798 | 37 | 0.495980 | 0.66 | 3.5e-08 | Click! |
| Nfkb2 | chr19_46304500_46305228 | 64 | 0.899573 | 0.65 | 9.4e-08 | Click! |
| Nfkb2 | chr19_46305872_46306101 | 12 | 0.837360 | 0.62 | 3.7e-07 | Click! |
| Nfkb2 | chr19_46304238_46304460 | 29 | 0.933051 | 0.54 | 2.1e-05 | Click! |
| Nfkb2 | chr19_46308814_46309463 | 3017 | 0.111979 | -0.52 | 3.9e-05 | Click! |
Activity of the Nfkb2 motif across conditions
Conditions sorted by the z-value of the Nfkb2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
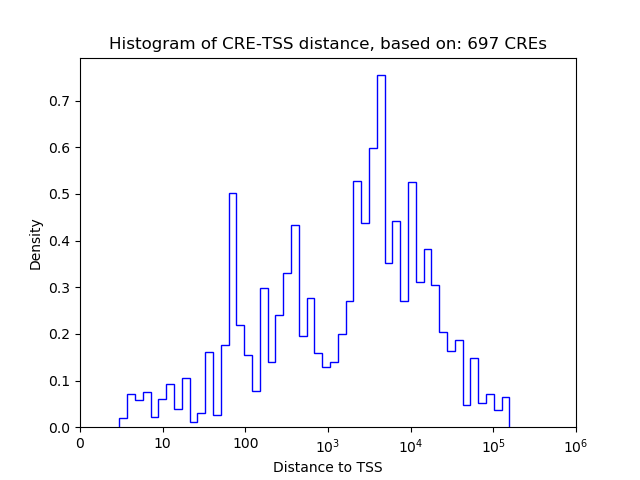
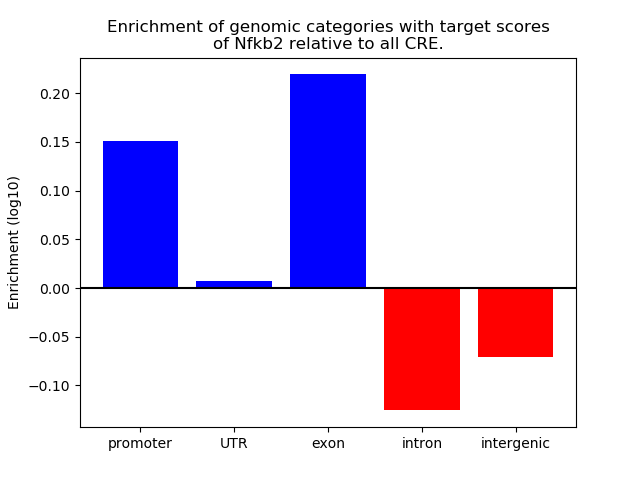
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_70865399_70865967 | 12.59 |
Kcnn1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
2425 |
0.11 |
| chr5_137530580_137532081 | 10.57 |
Gnb2 |
guanine nucleotide binding protein (G protein), beta 2 |
33 |
0.9 |
| chr17_29213743_29214236 | 10.29 |
Gm50054 |
predicted gene, 50054 |
9307 |
0.11 |
| chr5_137786077_137787112 | 9.89 |
Mepce |
methylphosphate capping enzyme |
69 |
0.92 |
| chr11_88068171_88069196 | 7.91 |
Vezf1 |
vascular endothelial zinc finger 1 |
404 |
0.76 |
| chr7_4739300_4740219 | 7.64 |
Kmt5c |
lysine methyltransferase 5C |
356 |
0.63 |
| chr15_98608664_98610204 | 6.69 |
Adcy6 |
adenylate cyclase 6 |
598 |
0.55 |
| chr19_46304500_46305228 | 6.13 |
Nfkb2 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
64 |
0.9 |
| chr11_102364651_102365006 | 5.34 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
419 |
0.72 |
| chr9_107312801_107313233 | 5.25 |
Gm17041 |
predicted gene 17041 |
11179 |
0.09 |
| chr9_111057150_111057866 | 5.12 |
Ccrl2 |
chemokine (C-C motif) receptor-like 2 |
11 |
0.95 |
| chr15_28025052_28026538 | 5.03 |
Trio |
triple functional domain (PTPRF interacting) |
53 |
0.98 |
| chr9_20804943_20805094 | 4.86 |
Col5a3 |
collagen, type V, alpha 3 |
10049 |
0.11 |
| chr9_5308573_5308892 | 4.63 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
96 |
0.97 |
| chr7_29511662_29511933 | 4.48 |
Sipa1l3 |
signal-induced proliferation-associated 1 like 3 |
6337 |
0.16 |
| chr7_116503804_116504452 | 4.40 |
Nucb2 |
nucleobindin 2 |
241 |
0.9 |
| chr2_38357064_38357839 | 4.37 |
Lhx2 |
LIM homeobox protein 2 |
3649 |
0.17 |
| chr19_5692044_5692416 | 4.36 |
Map3k11 |
mitogen-activated protein kinase kinase kinase 11 |
665 |
0.39 |
| chr3_51340298_51341882 | 4.36 |
Elf2 |
E74-like factor 2 |
427 |
0.74 |
| chr11_103175050_103175255 | 4.29 |
Fmnl1 |
formin-like 1 |
3839 |
0.14 |
| chr15_103014087_103015222 | 4.29 |
Mir615 |
microRNA 615 |
256 |
0.74 |
| chr6_33892312_33892463 | 4.26 |
Exoc4 |
exocyst complex component 4 |
30287 |
0.22 |
| chr8_122284945_122285186 | 4.24 |
Zfpm1 |
zinc finger protein, multitype 1 |
2924 |
0.21 |
| chr3_89898945_89899452 | 4.15 |
Gm42809 |
predicted gene 42809 |
13718 |
0.11 |
| chr11_69096542_69097348 | 4.09 |
Per1 |
period circadian clock 1 |
1728 |
0.15 |
| chr6_136938860_136939011 | 4.04 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
656 |
0.62 |
| chr2_170205883_170206176 | 4.03 |
Zfp217 |
zinc finger protein 217 |
57926 |
0.13 |
| chr7_47022714_47022865 | 3.99 |
Gm32031 |
predicted gene, 32031 |
14244 |
0.08 |
| chr12_113145608_113147138 | 3.97 |
Crip1 |
cysteine-rich protein 1 (intestinal) |
57 |
0.95 |
| chr9_70934534_70934685 | 3.95 |
Lipc |
lipase, hepatic |
4 |
0.98 |
| chr2_158305932_158306715 | 3.92 |
Lbp |
lipopolysaccharide binding protein |
170 |
0.92 |
| chr17_29394208_29394359 | 3.86 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
17782 |
0.11 |
| chr9_31602923_31603085 | 3.79 |
Gm18226 |
predicted gene, 18226 |
34758 |
0.13 |
| chr13_55210517_55212032 | 3.78 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
267 |
0.89 |
| chr2_162991719_162991888 | 3.78 |
Sgk2 |
serum/glucocorticoid regulated kinase 2 |
4301 |
0.15 |
| chr11_77785338_77785489 | 3.76 |
Gm10277 |
predicted gene 10277 |
2334 |
0.23 |
| chr11_53818950_53819101 | 3.67 |
Gm12216 |
predicted gene 12216 |
23150 |
0.11 |
| chr3_89136366_89136871 | 3.55 |
Pklr |
pyruvate kinase liver and red blood cell |
5 |
0.94 |
| chr2_73312496_73313091 | 3.54 |
Cir1 |
corepressor interacting with RBPJ, 1 |
92 |
0.53 |
| chr15_83215718_83216148 | 3.52 |
A4galt |
alpha 1,4-galactosyltransferase |
35796 |
0.09 |
| chr11_120304745_120305917 | 3.50 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
15538 |
0.09 |
| chr2_3418615_3419281 | 3.49 |
Meig1 |
meiosis expressed gene 1 |
71 |
0.96 |
| chr12_76883959_76884141 | 3.48 |
Fntb |
farnesyltransferase, CAAX box, beta |
3008 |
0.25 |
| chr9_90263012_90263305 | 3.46 |
Tbc1d2b |
TBC1 domain family, member 2B |
7231 |
0.18 |
| chr17_34000159_34000502 | 3.41 |
H2-K1 |
histocompatibility 2, K1, K region |
17 |
0.9 |
| chr11_49755060_49755211 | 3.39 |
Gm23813 |
predicted gene, 23813 |
16698 |
0.12 |
| chr5_28500848_28501098 | 3.39 |
Shh |
sonic hedgehog |
33717 |
0.14 |
| chr12_54369308_54369459 | 3.37 |
Gm7550 |
predicted gene 7550 |
21935 |
0.14 |
| chr10_98684197_98684591 | 3.36 |
Gm5427 |
predicted gene 5427 |
15316 |
0.28 |
| chr11_98766610_98767640 | 3.34 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3301 |
0.13 |
| chr10_80032799_80032976 | 3.34 |
Polr2e |
polymerase (RNA) II (DNA directed) polypeptide E |
4298 |
0.09 |
| chr18_32818614_32818817 | 3.30 |
Tslp |
thymic stromal lymphopoietin |
1855 |
0.27 |
| chr17_24878389_24878540 | 3.20 |
Igfals |
insulin-like growth factor binding protein, acid labile subunit |
206 |
0.84 |
| chr11_69946375_69946696 | 3.20 |
Slc2a4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
1439 |
0.15 |
| chr13_119619864_119620064 | 3.18 |
Ccl28 |
chemokine (C-C motif) ligand 28 |
3855 |
0.15 |
| chr19_5841346_5841905 | 3.17 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
3634 |
0.09 |
| chr4_16168525_16168676 | 3.17 |
A530072M11Rik |
RIKEN cDNA gene A530072M11 |
4490 |
0.19 |
| chr12_54317230_54317465 | 3.14 |
Gm47552 |
predicted gene, 47552 |
332 |
0.89 |
| chr17_15375662_15377138 | 3.14 |
Dll1 |
delta like canonical Notch ligand 1 |
424 |
0.8 |
| chr4_152345283_152345464 | 3.12 |
Chd5 |
chromodomain helicase DNA binding protein 5 |
3778 |
0.14 |
| chr3_89831199_89831385 | 3.09 |
She |
src homology 2 domain-containing transforming protein E |
78 |
0.95 |
| chr5_113985083_113985238 | 3.07 |
Ssh1 |
slingshot protein phosphatase 1 |
4583 |
0.15 |
| chr17_35218575_35218838 | 3.03 |
Gm16181 |
predicted gene 16181 |
2128 |
0.08 |
| chr11_50213339_50213741 | 3.02 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2650 |
0.15 |
| chr12_31080980_31081131 | 3.02 |
Fam110c |
family with sequence similarity 110, member C |
7194 |
0.13 |
| chr19_8984523_8984940 | 2.99 |
Ahnak |
AHNAK nucleoprotein (desmoyokin) |
4553 |
0.1 |
| chr2_75021769_75021920 | 2.99 |
n-R5s198 |
nuclear encoded rRNA 5S 198 |
86330 |
0.07 |
| chr14_8244059_8244558 | 2.92 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
2095 |
0.32 |
| chr17_26414662_26415402 | 2.91 |
Neurl1b |
neuralized E3 ubiquitin protein ligase 1B |
67 |
0.97 |
| chr3_95689670_95689851 | 2.91 |
Adamtsl4 |
ADAMTS-like 4 |
1843 |
0.21 |
| chr4_108317802_108317953 | 2.90 |
Gm12740 |
predicted gene 12740 |
4461 |
0.14 |
| chr13_37760115_37760291 | 2.83 |
Gm31600 |
predicted gene, 31600 |
1545 |
0.32 |
| chr13_93989303_93989588 | 2.82 |
Gm47216 |
predicted gene, 47216 |
2323 |
0.29 |
| chr6_42376343_42376494 | 2.76 |
2010310C07Rik |
RIKEN cDNA 2010310C07 gene |
1538 |
0.2 |
| chr19_55643252_55643620 | 2.71 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
98384 |
0.08 |
| chr7_141628110_141628282 | 2.70 |
Mir7063 |
microRNA 7063 |
7495 |
0.13 |
| chr7_120732949_120733317 | 2.65 |
Mosmo |
modulator of smoothened |
4809 |
0.13 |
| chr2_50964143_50964317 | 2.63 |
Gm13498 |
predicted gene 13498 |
54546 |
0.16 |
| chr16_95728896_95729375 | 2.61 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
11401 |
0.18 |
| chr9_72132750_72133422 | 2.61 |
Gm7863 |
predicted gene 7863 |
4499 |
0.13 |
| chr15_73521429_73521588 | 2.61 |
Dennd3 |
DENN/MADD domain containing 3 |
2626 |
0.26 |
| chr8_91395853_91396004 | 2.61 |
Gm45427 |
predicted gene 45427 |
3983 |
0.18 |
| chr3_105910590_105910826 | 2.60 |
Tmigd3 |
transmembrane and immunoglobulin domain containing 3 |
3428 |
0.14 |
| chr5_139812480_139812631 | 2.59 |
Tmem184a |
transmembrane protein 184a |
688 |
0.57 |
| chr9_44734596_44735576 | 2.59 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
112 |
0.91 |
| chr15_12479635_12479786 | 2.58 |
Pdzd2 |
PDZ domain containing 2 |
13849 |
0.22 |
| chr13_21936965_21937136 | 2.58 |
Zfp184 |
zinc finger protein 184 (Kruppel-like) |
8044 |
0.06 |
| chr1_133076912_133077418 | 2.57 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
8586 |
0.15 |
| chr10_76680967_76681118 | 2.56 |
Gm35608 |
predicted gene, 35608 |
3390 |
0.21 |
| chr11_95040343_95041272 | 2.54 |
Pdk2 |
pyruvate dehydrogenase kinase, isoenzyme 2 |
68 |
0.95 |
| chr4_63625447_63625664 | 2.51 |
Tex48 |
testis expressed 48 |
3127 |
0.16 |
| chr4_86873964_86874819 | 2.49 |
Acer2 |
alkaline ceramidase 2 |
5 |
0.98 |
| chr7_110156726_110157216 | 2.46 |
1600010M07Rik |
RIKEN cDNA 1600010M07 gene |
5769 |
0.16 |
| chr12_78206664_78206815 | 2.45 |
Gm6657 |
predicted gene 6657 |
5773 |
0.17 |
| chr14_54958317_54958468 | 2.42 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
485 |
0.52 |
| chr1_74007725_74008174 | 2.37 |
Tns1 |
tensin 1 |
6382 |
0.25 |
| chr10_69352241_69353323 | 2.36 |
Cdk1 |
cyclin-dependent kinase 1 |
121 |
0.96 |
| chr4_43964262_43964601 | 2.35 |
Glipr2 |
GLI pathogenesis-related 2 |
6739 |
0.14 |
| chr7_45053033_45053293 | 2.32 |
Prr12 |
proline rich 12 |
282 |
0.7 |
| chr17_47739478_47739853 | 2.31 |
Tfeb |
transcription factor EB |
762 |
0.49 |
| chr15_59728082_59728625 | 2.31 |
Gm20150 |
predicted gene, 20150 |
56240 |
0.12 |
| chr14_63899644_63899795 | 2.30 |
Pinx1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
50 |
0.98 |
| chr9_56871091_56871530 | 2.29 |
Cspg4 |
chondroitin sulfate proteoglycan 4 |
6277 |
0.13 |
| chr6_120579566_120580923 | 2.28 |
Gm44124 |
predicted gene, 44124 |
68 |
0.96 |
| chr18_60737123_60737515 | 2.26 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4169 |
0.18 |
| chr7_68709280_68709469 | 2.26 |
Gm44692 |
predicted gene 44692 |
17093 |
0.21 |
| chr1_43403457_43403608 | 2.24 |
Gm29041 |
predicted gene 29041 |
6052 |
0.23 |
| chr11_116220426_116220577 | 2.23 |
Cdk3 |
cyclin-dependent kinase 3 |
3961 |
0.11 |
| chr2_25095362_25095586 | 2.20 |
Noxa1 |
NADPH oxidase activator 1 |
325 |
0.75 |
| chr7_19822671_19822993 | 2.19 |
Bcl3 |
B cell leukemia/lymphoma 3 |
62 |
0.93 |
| chr7_136458176_136458476 | 2.19 |
Gm36849 |
predicted gene, 36849 |
104962 |
0.07 |
| chr8_93172146_93172297 | 2.18 |
Ces1d |
carboxylesterase 1D |
2246 |
0.22 |
| chr11_23770360_23771742 | 2.17 |
Rel |
reticuloendotheliosis oncogene |
81 |
0.97 |
| chr17_84146012_84146259 | 2.17 |
Gm19696 |
predicted gene, 19696 |
6294 |
0.18 |
| chr1_36096072_36096380 | 2.15 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
5464 |
0.15 |
| chr13_111210776_111210955 | 2.14 |
3110015C05Rik |
RIKEN cDNA 3110015C05 gene |
18566 |
0.21 |
| chr2_153528510_153529333 | 2.14 |
Nol4l |
nucleolar protein 4-like |
1050 |
0.54 |
| chr15_34046405_34047185 | 2.13 |
Gm49091 |
predicted gene, 49091 |
6323 |
0.17 |
| chr9_116091571_116091722 | 2.11 |
Gm9385 |
predicted pseudogene 9385 |
50582 |
0.13 |
| chr5_31095401_31095736 | 2.11 |
Gm9924 |
predicted gene 9924 |
582 |
0.41 |
| chr4_63233648_63233836 | 2.10 |
Col27a1 |
collagen, type XXVII, alpha 1 |
544 |
0.75 |
| chr4_154217759_154217994 | 2.08 |
Gm13132 |
predicted gene 13132 |
2665 |
0.22 |
| chr5_77386860_77387024 | 2.06 |
Gm34648 |
predicted gene, 34648 |
343 |
0.85 |
| chr5_147748059_147748210 | 2.04 |
Gm43156 |
predicted gene 43156 |
2618 |
0.28 |
| chr10_60081188_60081369 | 2.04 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
24941 |
0.16 |
| chr5_137482389_137482638 | 2.02 |
Epo |
erythropoietin |
3303 |
0.1 |
| chr14_25749259_25749410 | 2.01 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
19705 |
0.13 |
| chr1_89041400_89041551 | 1.96 |
Gm18699 |
predicted gene, 18699 |
7499 |
0.19 |
| chr7_29243253_29243404 | 1.96 |
Yif1b |
Yip1 interacting factor homolog B (S. cerevisiae) |
4801 |
0.1 |
| chr10_63023265_63024505 | 1.95 |
Hnrnph3 |
heterogeneous nuclear ribonucleoprotein H3 |
9 |
0.86 |
| chr8_122702350_122702501 | 1.94 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
3316 |
0.13 |
| chr19_45114191_45114342 | 1.93 |
Gm35867 |
predicted gene, 35867 |
11025 |
0.14 |
| chr2_32315786_32315983 | 1.92 |
Dnm1 |
dynamin 1 |
1447 |
0.19 |
| chr9_120492056_120493362 | 1.92 |
D830035M03Rik |
RIKEN cDNA D830035M03 gene |
162 |
0.57 |
| chr18_51796497_51796650 | 1.92 |
Gm4950 |
predicted pseudogene 4950 |
69308 |
0.13 |
| chr9_52047751_52048337 | 1.91 |
Rdx |
radixin |
71 |
0.97 |
| chr5_120588908_120589354 | 1.90 |
Iqcd |
IQ motif containing D |
108 |
0.82 |
| chr7_24459503_24459679 | 1.90 |
Plaur |
plasminogen activator, urokinase receptor |
2893 |
0.12 |
| chr15_73753083_73753263 | 1.89 |
Ptp4a3 |
protein tyrosine phosphatase 4a3 |
4799 |
0.19 |
| chr12_110219876_110220027 | 1.89 |
Gm40576 |
predicted gene, 40576 |
16411 |
0.11 |
| chr4_152115745_152116129 | 1.87 |
Tnfrsf25 |
tumor necrosis factor receptor superfamily, member 25 |
3 |
0.96 |
| chr4_133185373_133185552 | 1.87 |
Wasf2 |
WAS protein family, member 2 |
3556 |
0.16 |
| chr2_132923726_132924027 | 1.86 |
Fermt1 |
fermitin family member 1 |
18087 |
0.14 |
| chr5_105732415_105732763 | 1.85 |
Lrrc8d |
leucine rich repeat containing 8D |
359 |
0.89 |
| chr7_46724003_46724164 | 1.85 |
Saa3 |
serum amyloid A 3 |
8383 |
0.09 |
| chr11_78848553_78848704 | 1.85 |
Lyrm9 |
LYR motif containing 9 |
21999 |
0.15 |
| chr2_153083371_153083522 | 1.85 |
Ccm2l |
cerebral cavernous malformation 2-like |
17444 |
0.13 |
| chr14_60736854_60737045 | 1.84 |
Spata13 |
spermatogenesis associated 13 |
4043 |
0.23 |
| chr2_167755309_167755460 | 1.84 |
Gm14321 |
predicted gene 14321 |
22083 |
0.12 |
| chr4_155233625_155234685 | 1.83 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
11563 |
0.16 |
| chr3_27182778_27183563 | 1.83 |
Nceh1 |
neutral cholesterol ester hydrolase 1 |
166 |
0.95 |
| chr4_135312755_135313155 | 1.81 |
Gm12982 |
predicted gene 12982 |
4985 |
0.13 |
| chr7_141103278_141103452 | 1.81 |
Ano9 |
anoctamin 9 |
689 |
0.42 |
| chr5_121848352_121848558 | 1.79 |
4933437G19Rik |
RIKEN cDNA 4933437G19 gene |
389 |
0.53 |
| chr7_19817396_19818051 | 1.78 |
Gm16174 |
predicted gene 16174 |
1170 |
0.22 |
| chr6_5156932_5157313 | 1.78 |
Pon1 |
paraoxonase 1 |
36641 |
0.14 |
| chr1_74295108_74296112 | 1.78 |
Gm29358 |
predicted gene 29358 |
7 |
0.55 |
| chr7_101301390_101302659 | 1.77 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
3 |
0.96 |
| chr17_59382519_59382795 | 1.75 |
Gm23769 |
predicted gene, 23769 |
156814 |
0.04 |
| chr4_135686362_135686542 | 1.74 |
Ifnlr1 |
interferon lambda receptor 1 |
165 |
0.94 |
| chr4_115094117_115094298 | 1.74 |
Pdzk1ip1 |
PDZK1 interacting protein 1 |
3275 |
0.19 |
| chr1_160043758_160043909 | 1.74 |
Gm37294 |
predicted gene, 37294 |
498 |
0.48 |
| chr7_90023149_90023335 | 1.73 |
E230029C05Rik |
RIKEN cDNA E230029C05 gene |
5917 |
0.14 |
| chr15_73668190_73668541 | 1.69 |
1700010B13Rik |
RIKEN cDNA 1700010B13 gene |
22491 |
0.14 |
| chr14_21746972_21747123 | 1.69 |
Dusp13 |
dual specificity phosphatase 13 |
562 |
0.67 |
| chr14_65374130_65374281 | 1.67 |
Zfp395 |
zinc finger protein 395 |
1188 |
0.46 |
| chr18_32432365_32432579 | 1.67 |
Bin1 |
bridging integrator 1 |
4438 |
0.19 |
| chr2_167589238_167589449 | 1.66 |
Gm11475 |
predicted gene 11475 |
2052 |
0.21 |
| chr13_21924698_21924984 | 1.66 |
Gm44456 |
predicted gene, 44456 |
175 |
0.85 |
| chr4_129951627_129951778 | 1.66 |
Spocd1 |
SPOC domain containing 1 |
4155 |
0.15 |
| chr10_78136004_78136160 | 1.66 |
Gm47922 |
predicted gene, 47922 |
2015 |
0.23 |
| chr4_145319338_145319489 | 1.65 |
Tnfrsf8 |
tumor necrosis factor receptor superfamily, member 8 |
4249 |
0.22 |
| chr2_84865643_84866287 | 1.64 |
Rtn4rl2 |
reticulon 4 receptor-like 2 |
20511 |
0.1 |
| chr11_98394155_98394317 | 1.63 |
Pgap3 |
post-GPI attachment to proteins 3 |
2164 |
0.14 |
| chr3_36612816_36613750 | 1.61 |
Bbs7 |
Bardet-Biedl syndrome 7 (human) |
2 |
0.97 |
| chr14_30625908_30626847 | 1.61 |
Prkcd |
protein kinase C, delta |
167 |
0.94 |
| chr9_45803149_45803345 | 1.60 |
Cep164 |
centrosomal protein 164 |
9 |
0.96 |
| chr1_184173090_184173241 | 1.60 |
Dusp10 |
dual specificity phosphatase 10 |
138784 |
0.04 |
| chr4_125127322_125128114 | 1.57 |
Zc3h12a |
zinc finger CCCH type containing 12A |
122 |
0.95 |
| chr17_33918604_33919342 | 1.56 |
Tapbp |
TAP binding protein |
359 |
0.55 |
| chr10_77110050_77110376 | 1.56 |
Col18a1 |
collagen, type XVIII, alpha 1 |
3492 |
0.22 |
| chr9_61946662_61947035 | 1.56 |
Kif23 |
kinesin family member 23 |
74 |
0.97 |
| chr7_98558638_98558867 | 1.56 |
A630091E08Rik |
RIKEN cDNA A630091E08 gene |
4238 |
0.19 |
| chr1_182281988_182282369 | 1.55 |
Degs1 |
delta(4)-desaturase, sphingolipid 1 |
46 |
0.97 |
| chr5_96622772_96622943 | 1.54 |
Fras1 |
Fraser extracellular matrix complex subunit 1 |
5276 |
0.28 |
| chr1_130832611_130832799 | 1.54 |
Pigr |
polymeric immunoglobulin receptor |
3695 |
0.15 |
| chr11_50212819_50213300 | 1.53 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2169 |
0.17 |
| chr11_115839569_115839994 | 1.53 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
5458 |
0.11 |
| chr10_19937094_19937381 | 1.51 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
2014 |
0.34 |
| chr15_78763229_78763555 | 1.51 |
Mfng |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
3772 |
0.16 |
| chr15_93005718_93005899 | 1.51 |
Gm30085 |
predicted gene, 30085 |
3476 |
0.27 |
| chr12_111882999_111883158 | 1.51 |
Gm19195 |
predicted gene, 19195 |
21942 |
0.11 |
| chr11_88848741_88848959 | 1.51 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
2653 |
0.21 |
| chr9_59952350_59952501 | 1.51 |
Gm7616 |
predicted gene 7616 |
1140 |
0.35 |
| chr14_65149467_65150281 | 1.51 |
Extl3 |
exostosin-like glycosyltransferase 3 |
19 |
0.97 |
| chr5_43948006_43948538 | 1.51 |
Gm43183 |
predicted gene 43183 |
156 |
0.73 |
| chr4_125640472_125640832 | 1.49 |
Mir692-2 |
microRNA 692-2 |
135903 |
0.04 |
| chr1_191278513_191278664 | 1.49 |
Gm37168 |
predicted gene, 37168 |
11410 |
0.13 |
| chr12_8443374_8443541 | 1.49 |
Gm48076 |
predicted gene, 48076 |
11461 |
0.16 |
| chr15_61168959_61169125 | 1.48 |
Gm38563 |
predicted gene, 38563 |
11173 |
0.22 |
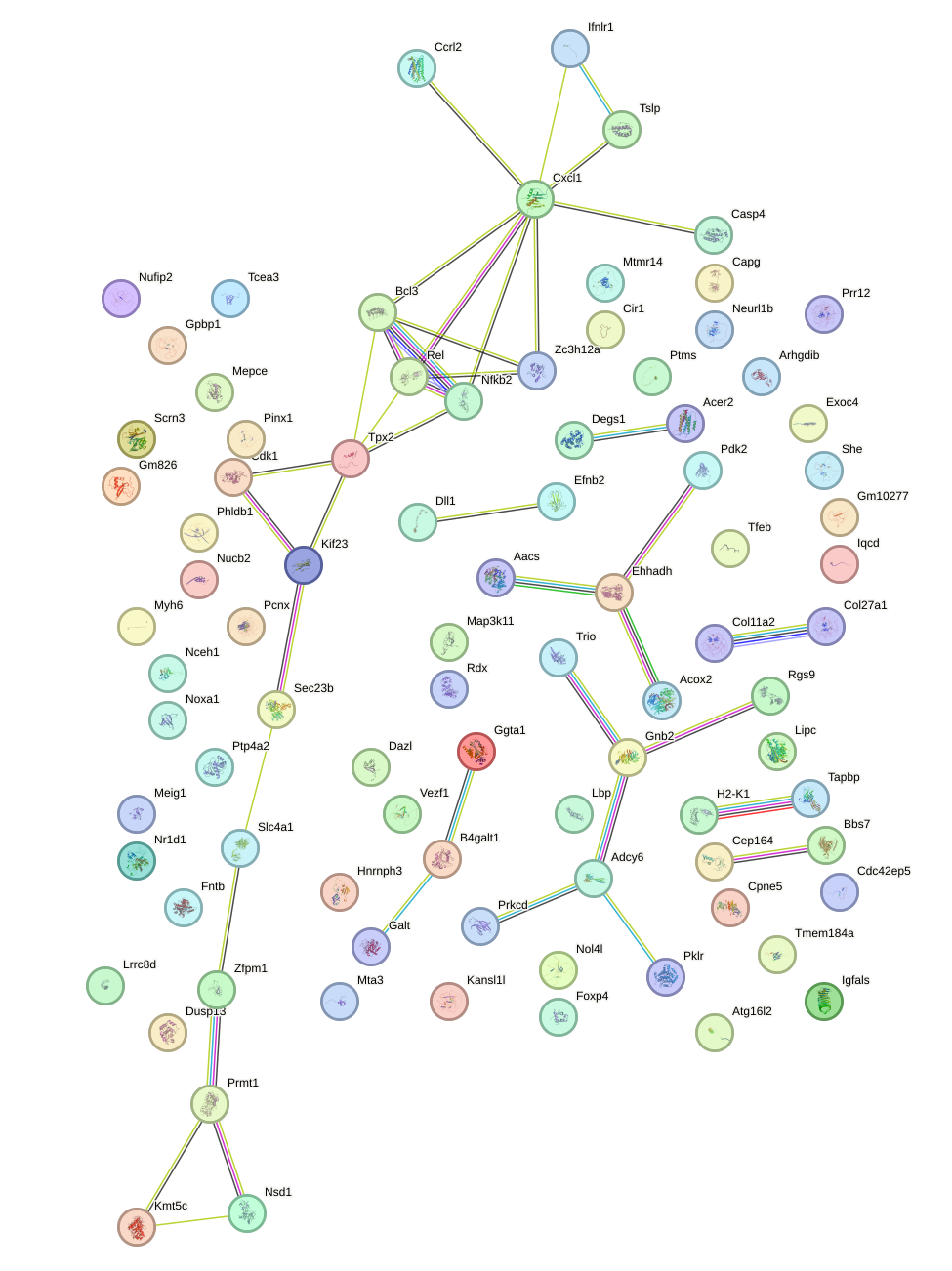
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 1.3 | 4.0 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 1.0 | 4.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.9 | 3.7 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.9 | 2.7 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.8 | 2.4 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.8 | 2.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.8 | 2.3 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.7 | 2.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.7 | 2.9 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.7 | 4.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.7 | 2.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.7 | 3.9 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.6 | 2.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.6 | 2.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.6 | 1.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.6 | 3.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 3.7 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.5 | 4.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.5 | 4.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.4 | 2.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.4 | 1.8 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 1.3 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.4 | 1.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.4 | 1.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 2.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.4 | 1.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 5.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.4 | 1.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.4 | 1.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.3 | 1.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 1.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 2.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 | 1.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.3 | 0.8 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 1.2 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.2 | 4.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.2 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.2 | 1.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.5 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.2 | 0.6 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 1.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.2 | 0.3 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.2 | 0.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 0.3 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.2 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 0.6 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 0.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.2 | 0.5 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 0.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.2 | 0.5 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.9 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.4 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 1.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.6 | GO:0009445 | putrescine metabolic process(GO:0009445) |
| 0.1 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 1.0 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 0.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.2 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.3 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 0.7 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 5.8 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 1.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.3 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 2.0 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.2 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.1 | 0.9 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 1.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.2 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.1 | 0.3 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.6 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.3 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.3 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 0.4 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.4 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.1 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.9 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.3 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.1 | 0.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.4 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.2 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.1 | 0.4 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.2 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.1 | 0.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.2 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.5 | GO:0032966 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.1 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.4 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.2 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 0.3 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.2 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.1 | GO:0072173 | metanephric tubule morphogenesis(GO:0072173) |
| 0.1 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.1 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.1 | 0.2 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.1 | 0.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.5 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.9 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 1.8 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.1 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.4 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 1.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.0 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.4 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 2.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.9 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.3 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 1.1 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.1 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 3.8 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.0 | GO:0048819 | regulation of hair follicle maturation(GO:0048819) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.0 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.0 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0001768 | establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.0 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0048103 | somatic stem cell division(GO:0048103) |
| 0.0 | 0.0 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:1901679 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.0 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.4 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.0 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0090500 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.7 | GO:0071356 | cellular response to tumor necrosis factor(GO:0071356) |
| 0.0 | 0.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.0 | GO:0034443 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.0 | GO:0061085 | regulation of histone H3-K27 methylation(GO:0061085) |
| 0.0 | 3.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.0 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.0 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.0 | 0.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.2 | GO:0042384 | cilium assembly(GO:0042384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 10.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.1 | 4.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.8 | 2.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.7 | 2.7 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 1.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.4 | 2.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 2.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 10.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 4.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 3.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.7 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 3.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 4.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.0 | GO:0005818 | aster(GO:0005818) |
| 0.2 | 1.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.1 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 3.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 2.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 4.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.3 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0031231 | intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 1.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.0 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.8 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 3.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.7 | 3.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.7 | 2.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.6 | 3.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 3.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 1.5 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.5 | 2.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 1.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.4 | 1.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.4 | 5.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 2.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 2.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 9.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 0.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 0.9 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 3.5 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.2 | 2.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 1.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 3.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 0.6 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 0.6 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 3.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.4 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 4.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 9.2 | GO:0080012 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.1 | 2.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.9 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 3.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.3 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0034810 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.5 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 3.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0042557 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 2.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.3 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.7 | GO:0052768 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.0 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0035586 | purinergic receptor activity(GO:0035586) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0033558 | protein deacetylase activity(GO:0033558) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 3.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.2 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.2 | GO:0052744 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0001618 | virus receptor activity(GO:0001618) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 6.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 3.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 3.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 4.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 6.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 7.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 2.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 4.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 5.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 2.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 4.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 5.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.2 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.1 | 1.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 2.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |