Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
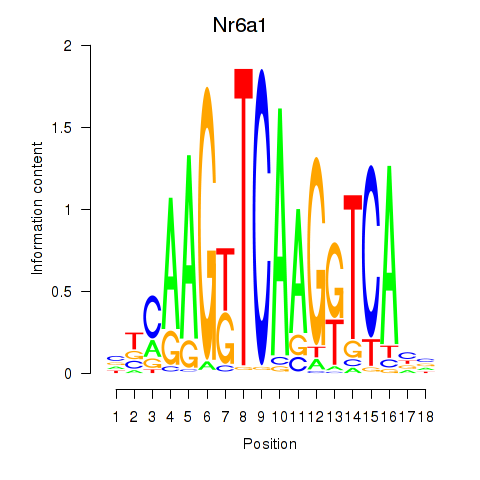
Results for Nr6a1
Z-value: 0.98
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.7 | Nr6a1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Nr6a1 | chr2_38739566_38739717 | 823 | 0.485625 | 0.49 | 1.3e-04 | Click! |
| Nr6a1 | chr2_38784488_38784649 | 48 | 0.957925 | -0.20 | 1.5e-01 | Click! |
| Nr6a1 | chr2_38925778_38926094 | 281 | 0.838462 | 0.05 | 7.4e-01 | Click! |
| Nr6a1 | chr2_38926098_38926539 | 43 | 0.954949 | 0.04 | 7.6e-01 | Click! |
| Nr6a1 | chr2_38785249_38785400 | 213 | 0.893798 | -0.04 | 7.6e-01 | Click! |
Activity of the Nr6a1 motif across conditions
Conditions sorted by the z-value of the Nr6a1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
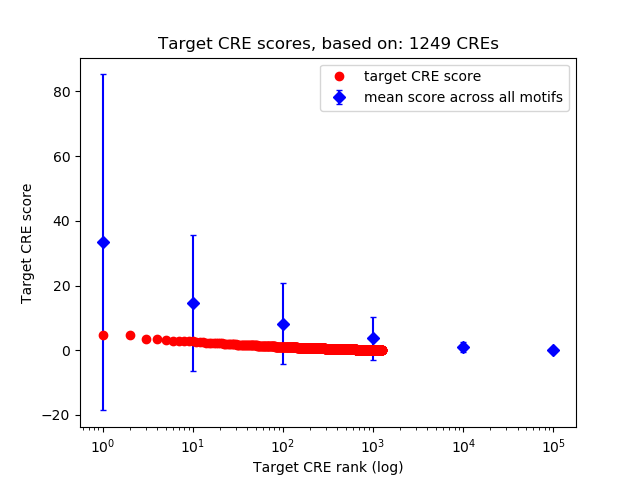
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_110050502_110051534 | 4.71 |
Dmrta2 |
doublesex and mab-3 related transcription factor like family A2 |
72965 |
0.11 |
| chr9_124439906_124440949 | 4.68 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
441 |
0.79 |
| chr4_82304607_82304811 | 3.62 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
134701 |
0.05 |
| chr1_81520769_81521201 | 3.59 |
Gm37210 |
predicted gene, 37210 |
1356 |
0.55 |
| chr1_81588038_81588300 | 3.09 |
Gm6198 |
predicted gene 6198 |
30686 |
0.22 |
| chr4_98108669_98109025 | 2.98 |
Gm12691 |
predicted gene 12691 |
37752 |
0.2 |
| chr10_76732571_76732722 | 2.96 |
Gm48276 |
predicted gene, 48276 |
2373 |
0.25 |
| chr11_117336202_117336679 | 2.90 |
Septin9 |
septin 9 |
4012 |
0.23 |
| chr6_119409613_119410171 | 2.88 |
Adipor2 |
adiponectin receptor 2 |
7583 |
0.21 |
| chr12_3236518_3237725 | 2.77 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
510 |
0.74 |
| chr13_40994999_40995221 | 2.54 |
Pak1ip1 |
PAK1 interacting protein 1 |
5913 |
0.12 |
| chrX_60147349_60147500 | 2.43 |
Mcf2 |
mcf.2 transforming sequence |
219 |
0.95 |
| chr4_82505225_82505396 | 2.43 |
Nfib |
nuclear factor I/B |
0 |
0.98 |
| chr10_11044838_11044994 | 2.38 |
Gm48406 |
predicted gene, 48406 |
9000 |
0.23 |
| chr17_49472723_49472906 | 2.30 |
Mocs1 |
molybdenum cofactor synthesis 1 |
20530 |
0.2 |
| chr18_11234055_11234212 | 2.19 |
Gata6 |
GATA binding protein 6 |
175086 |
0.03 |
| chr12_95695278_95696175 | 2.13 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
369 |
0.86 |
| chr11_55022601_55023592 | 2.12 |
Anxa6 |
annexin A6 |
10319 |
0.16 |
| chr13_63230991_63231626 | 2.10 |
Aopep |
aminopeptidase O |
8721 |
0.11 |
| chr12_108003961_108004329 | 2.10 |
Bcl11b |
B cell leukemia/lymphoma 11B |
543 |
0.85 |
| chr4_119815571_119815722 | 2.09 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
1151 |
0.61 |
| chr19_10071333_10071499 | 2.06 |
Fads2 |
fatty acid desaturase 2 |
229 |
0.9 |
| chr16_63747767_63748162 | 2.02 |
Gm22769 |
predicted gene, 22769 |
430 |
0.91 |
| chr7_16119631_16120684 | 1.99 |
Kptn |
kaptin |
62 |
0.96 |
| chr13_69482238_69482460 | 1.93 |
Gm48676 |
predicted gene, 48676 |
31651 |
0.13 |
| chr7_79535477_79536145 | 1.90 |
Gm35040 |
predicted gene, 35040 |
232 |
0.85 |
| chr17_74879880_74880031 | 1.86 |
Ttc27 |
tetratricopeptide repeat domain 27 |
18034 |
0.17 |
| chr2_58787447_58787599 | 1.81 |
Upp2 |
uridine phosphorylase 2 |
22198 |
0.18 |
| chr1_90093233_90093384 | 1.79 |
Iqca |
IQ motif containing with AAA domain |
3164 |
0.29 |
| chr9_58355790_58355941 | 1.77 |
Tbc1d21 |
TBC1 domain family, member 21 |
14504 |
0.15 |
| chr4_32196712_32196887 | 1.73 |
Gm11929 |
predicted gene 11929 |
10349 |
0.2 |
| chr5_114175447_114176205 | 1.72 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
80 |
0.96 |
| chr17_56809917_56810952 | 1.72 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
20505 |
0.11 |
| chr10_110927477_110927905 | 1.70 |
Csrp2 |
cysteine and glycine-rich protein 2 |
3510 |
0.19 |
| chr6_119487122_119487339 | 1.68 |
Fbxl14 |
F-box and leucine-rich repeat protein 14 |
7562 |
0.21 |
| chr1_57846936_57847100 | 1.67 |
Spats2l |
spermatogenesis associated, serine-rich 2-like |
1447 |
0.5 |
| chr10_107752299_107752727 | 1.66 |
Ptprq |
protein tyrosine phosphatase, receptor type, Q |
32462 |
0.23 |
| chr17_25462548_25463080 | 1.65 |
Tekt4 |
tektin 4 |
8199 |
0.11 |
| chr8_102596538_102596950 | 1.60 |
Cdh11 |
cadherin 11 |
40575 |
0.15 |
| chr11_111215086_111215415 | 1.60 |
Gm11675 |
predicted gene 11675 |
60850 |
0.15 |
| chr3_9704898_9705332 | 1.58 |
Pag1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
113 |
0.97 |
| chr2_115660293_115660477 | 1.58 |
Mir1951 |
microRNA 1951 |
21660 |
0.23 |
| chr1_79858099_79859256 | 1.57 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
19 |
0.98 |
| chr6_134994369_134994520 | 1.56 |
Apold1 |
apolipoprotein L domain containing 1 |
12726 |
0.13 |
| chr11_120311318_120311784 | 1.56 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
21758 |
0.08 |
| chr8_10754564_10754760 | 1.51 |
Gm39129 |
predicted gene, 39129 |
15772 |
0.13 |
| chr4_141425494_141426465 | 1.49 |
Hspb7 |
heat shock protein family, member 7 (cardiovascular) |
5200 |
0.1 |
| chr6_93648670_93648869 | 1.49 |
Gm7812 |
predicted gene 7812 |
1456 |
0.55 |
| chr5_112825174_112825344 | 1.47 |
1700095B10Rik |
RIKEN cDNA 1700095B10 gene |
31927 |
0.16 |
| chr12_10017131_10017323 | 1.47 |
Gm5182 |
predicted gene 5182 |
134915 |
0.05 |
| chr4_76461421_76461659 | 1.46 |
Gm42303 |
predicted gene, 42303 |
10309 |
0.22 |
| chr16_89924625_89924841 | 1.45 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
5748 |
0.31 |
| chr15_71917480_71917631 | 1.45 |
Col22a1 |
collagen, type XXII, alpha 1 |
19939 |
0.24 |
| chr7_4516460_4516712 | 1.45 |
Tnnt1 |
troponin T1, skeletal, slow |
204 |
0.84 |
| chr13_39823948_39824404 | 1.43 |
A230103O09Rik |
RIKEN cDNA A230103O09 gene |
178 |
0.94 |
| chr1_75559272_75559776 | 1.40 |
Slc4a3 |
solute carrier family 4 (anion exchanger), member 3 |
7815 |
0.11 |
| chr17_56467426_56468397 | 1.40 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
5695 |
0.16 |
| chr8_126659831_126660263 | 1.36 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
66061 |
0.11 |
| chr17_88954148_88954333 | 1.34 |
Gm9407 |
predicted gene 9407 |
97618 |
0.08 |
| chr8_6456655_6456806 | 1.34 |
Gm44844 |
predicted gene 44844 |
18986 |
0.29 |
| chr4_116016940_116018214 | 1.33 |
Faah |
fatty acid amide hydrolase |
98 |
0.95 |
| chr14_32321782_32322361 | 1.33 |
Ogdhl |
oxoglutarate dehydrogenase-like |
52 |
0.96 |
| chr3_9833533_9834642 | 1.32 |
Pag1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
408 |
0.84 |
| chr9_95257001_95257349 | 1.29 |
Slc9a9 |
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
135489 |
0.04 |
| chr14_12716331_12716619 | 1.29 |
Gm48266 |
predicted gene, 48266 |
54075 |
0.13 |
| chr1_74054406_74054750 | 1.28 |
Tns1 |
tensin 1 |
17421 |
0.19 |
| chr1_73992293_73993185 | 1.27 |
Tns1 |
tensin 1 |
2633 |
0.33 |
| chr8_127438974_127439332 | 1.26 |
Pard3 |
par-3 family cell polarity regulator |
8593 |
0.3 |
| chr11_69559283_69560047 | 1.25 |
Efnb3 |
ephrin B3 |
540 |
0.54 |
| chr17_57276298_57276514 | 1.24 |
Vav1 |
vav 1 oncogene |
2694 |
0.18 |
| chr10_18468496_18468770 | 1.24 |
Nhsl1 |
NHS-like 1 |
1255 |
0.55 |
| chr5_125163793_125164061 | 1.24 |
Ncor2 |
nuclear receptor co-repressor 2 |
15126 |
0.21 |
| chr4_114676951_114677102 | 1.23 |
Gm28864 |
predicted gene 28864 |
3686 |
0.25 |
| chr1_194030707_194030867 | 1.23 |
Gm21362 |
predicted gene, 21362 |
163778 |
0.04 |
| chr6_114396692_114396879 | 1.22 |
Hrh1 |
histamine receptor H1 |
1151 |
0.61 |
| chr5_125216280_125216449 | 1.21 |
Ncor2 |
nuclear receptor co-repressor 2 |
37145 |
0.14 |
| chr2_90917239_90918445 | 1.21 |
Ptpmt1 |
protein tyrosine phosphatase, mitochondrial 1 |
15 |
0.71 |
| chr17_80803081_80803318 | 1.19 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
74714 |
0.09 |
| chr8_34163517_34163668 | 1.18 |
Mir6395 |
microRNA 6395 |
3120 |
0.14 |
| chr13_55362078_55363351 | 1.15 |
Lman2 |
lectin, mannose-binding 2 |
69 |
0.95 |
| chr8_44708951_44709102 | 1.15 |
Gm26089 |
predicted gene, 26089 |
107193 |
0.07 |
| chr1_127352497_127352650 | 1.15 |
Gm23370 |
predicted gene, 23370 |
20760 |
0.15 |
| chr7_112222783_112223211 | 1.15 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
2859 |
0.36 |
| chr15_62230752_62231120 | 1.14 |
Pvt1 |
Pvt1 oncogene |
8333 |
0.25 |
| chr8_84712009_84712803 | 1.14 |
Nfix |
nuclear factor I/X |
1662 |
0.21 |
| chr13_51408491_51409234 | 1.13 |
S1pr3 |
sphingosine-1-phosphate receptor 3 |
223 |
0.94 |
| chr2_150645270_150645552 | 1.11 |
Acss1 |
acyl-CoA synthetase short-chain family member 1 |
7296 |
0.13 |
| chr3_115822572_115822723 | 1.11 |
Dph5 |
diphthamide biosynthesis 5 |
65190 |
0.08 |
| chr11_9048085_9048240 | 1.11 |
Gm11992 |
predicted gene 11992 |
432 |
0.67 |
| chr9_102692932_102693123 | 1.09 |
4930533D04Rik |
RIKEN cDNA 4930533D04 gene |
142 |
0.94 |
| chr13_53205183_53205803 | 1.09 |
Ror2 |
receptor tyrosine kinase-like orphan receptor 2 |
70165 |
0.11 |
| chr14_61327668_61327972 | 1.08 |
Gm5461 |
predicted gene 5461 |
2010 |
0.21 |
| chr2_56983190_56983369 | 1.08 |
Gm13527 |
predicted gene 13527 |
96469 |
0.08 |
| chr8_48224421_48225088 | 1.08 |
Gm32842 |
predicted gene, 32842 |
46231 |
0.16 |
| chr9_62436154_62436349 | 1.06 |
Coro2b |
coronin, actin binding protein, 2B |
7001 |
0.24 |
| chr16_77296088_77296239 | 1.06 |
Gm31258 |
predicted gene, 31258 |
9397 |
0.2 |
| chr3_127662316_127662467 | 1.04 |
Alpk1 |
alpha-kinase 1 |
23949 |
0.1 |
| chr10_116243379_116243665 | 1.04 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
32001 |
0.15 |
| chrX_71051287_71051495 | 1.04 |
Mamld1 |
mastermind-like domain containing 1 |
1135 |
0.58 |
| chr14_70530107_70530260 | 1.04 |
Lgi3 |
leucine-rich repeat LGI family, member 3 |
502 |
0.65 |
| chr18_15405119_15405460 | 1.03 |
Aqp4 |
aquaporin 4 |
1531 |
0.37 |
| chr7_68744740_68744917 | 1.01 |
Arrdc4 |
arrestin domain containing 4 |
4360 |
0.26 |
| chr1_176949345_176949641 | 1.01 |
Gm15423 |
predicted gene 15423 |
16782 |
0.13 |
| chr17_23960426_23960630 | 1.01 |
Sbpl |
spermine binding protein-like |
5254 |
0.08 |
| chr3_107517078_107517566 | 1.00 |
Slc6a17 |
solute carrier family 6 (neurotransmitter transporter), member 17 |
5 |
0.98 |
| chr10_79681206_79682337 | 1.00 |
Cdc34 |
cell division cycle 34 |
424 |
0.63 |
| chr11_108241078_108241308 | 1.00 |
Gm11655 |
predicted gene 11655 |
59343 |
0.12 |
| chr8_128025780_128025931 | 0.98 |
Mir21c |
microRNA 21c |
252370 |
0.02 |
| chr11_66487857_66488008 | 0.97 |
Shisa6 |
shisa family member 6 |
14882 |
0.27 |
| chr8_20384730_20385046 | 0.96 |
Gm7807 |
predicted gene 7807 |
3807 |
0.22 |
| chr10_34563895_34564051 | 0.96 |
Frk |
fyn-related kinase |
80441 |
0.1 |
| chr5_30920760_30922186 | 0.95 |
Khk |
ketohexokinase |
42 |
0.93 |
| chr11_118247752_118249318 | 0.95 |
Cyth1 |
cytohesin 1 |
25 |
0.97 |
| chr18_44754207_44754358 | 0.95 |
Mcc |
mutated in colorectal cancers |
29624 |
0.17 |
| chr2_114048115_114048266 | 0.95 |
Actc1 |
actin, alpha, cardiac muscle 1 |
4697 |
0.18 |
| chr12_54288052_54288203 | 0.94 |
Gm47552 |
predicted gene, 47552 |
28888 |
0.13 |
| chr8_128263663_128263945 | 0.94 |
Mir21c |
microRNA 21c |
14421 |
0.27 |
| chr5_151142402_151142622 | 0.94 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
14969 |
0.24 |
| chr12_11436805_11437388 | 0.92 |
Vsnl1 |
visinin-like 1 |
483 |
0.68 |
| chr7_66955436_66955938 | 0.92 |
Gm44755 |
predicted gene 44755 |
6231 |
0.22 |
| chr15_27630977_27631306 | 0.92 |
Otulin |
OTU deubiquitinase with linear linkage specificity |
448 |
0.79 |
| chr16_77239477_77239636 | 0.92 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
3237 |
0.3 |
| chr5_105793170_105793576 | 0.91 |
Rps15a-ps5 |
ribosomal protein S15A, pseudogene 5 |
4340 |
0.19 |
| chr3_83997060_83997321 | 0.91 |
Tmem131l |
transmembrane 131 like |
28962 |
0.2 |
| chr7_139516348_139516610 | 0.91 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
180 |
0.96 |
| chr12_104897006_104897472 | 0.91 |
Gm28875 |
predicted gene 28875 |
29450 |
0.13 |
| chr5_64580424_64580579 | 0.90 |
Gm42566 |
predicted gene 42566 |
15682 |
0.11 |
| chr16_34902354_34902568 | 0.90 |
Mylk |
myosin, light polypeptide kinase |
2206 |
0.34 |
| chr19_27607032_27607183 | 0.90 |
Gm50101 |
predicted gene, 50101 |
94599 |
0.08 |
| chr2_75000578_75000751 | 0.89 |
n-R5s198 |
nuclear encoded rRNA 5S 198 |
65150 |
0.09 |
| chr19_20567101_20567252 | 0.89 |
C730002L08Rik |
RIKEN cDNA C730002L08 gene |
14846 |
0.19 |
| chr2_38425074_38425256 | 0.88 |
Gm13589 |
predicted gene 13589 |
1163 |
0.43 |
| chr11_77875222_77875543 | 0.88 |
Pipox |
pipecolic acid oxidase |
7304 |
0.15 |
| chr2_17839635_17839891 | 0.88 |
Gm13323 |
predicted gene 13323 |
41779 |
0.17 |
| chr3_134105217_134105368 | 0.88 |
Gm26691 |
predicted gene, 26691 |
3227 |
0.26 |
| chr2_52050954_52051105 | 0.86 |
Tnfaip6 |
tumor necrosis factor alpha induced protein 6 |
13020 |
0.19 |
| chr8_10040892_10041043 | 0.86 |
Tnfsf13b |
tumor necrosis factor (ligand) superfamily, member 13b |
9649 |
0.16 |
| chr17_25433197_25433617 | 0.86 |
Cacna1h |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
129 |
0.93 |
| chr15_83037870_83038112 | 0.86 |
Nfam1 |
Nfat activating molecule with ITAM motif 1 |
4685 |
0.16 |
| chr6_51513838_51514067 | 0.85 |
Snx10 |
sorting nexin 10 |
9949 |
0.19 |
| chr10_83692278_83692464 | 0.85 |
1500009L16Rik |
RIKEN cDNA 1500009L16 gene |
30494 |
0.15 |
| chr14_84446496_84447465 | 0.84 |
Pcdh17 |
protocadherin 17 |
1527 |
0.47 |
| chr11_3132475_3133419 | 0.84 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
3060 |
0.17 |
| chr9_77496504_77496829 | 0.83 |
Lrrc1 |
leucine rich repeat containing 1 |
35778 |
0.13 |
| chr8_104602084_104602428 | 0.83 |
Pdp2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
10805 |
0.09 |
| chr8_20257716_20258061 | 0.83 |
6820431F20Rik |
RIKEN cDNA 6820431F20 gene |
25378 |
0.2 |
| chr12_15566567_15566816 | 0.83 |
Gm17915 |
predicted gene, 17915 |
14563 |
0.23 |
| chr3_28264071_28264435 | 0.83 |
Tnik |
TRAF2 and NCK interacting kinase |
610 |
0.76 |
| chr6_89008167_89008318 | 0.83 |
4933427D06Rik |
RIKEN cDNA 4933427D06 gene |
57559 |
0.1 |
| chr3_52685799_52685950 | 0.82 |
Gm22798 |
predicted gene, 22798 |
36881 |
0.16 |
| chr2_54215819_54215970 | 0.82 |
Gm13505 |
predicted gene 13505 |
24675 |
0.22 |
| chr3_107793294_107794069 | 0.81 |
Gm43233 |
predicted gene 43233 |
11825 |
0.12 |
| chr16_17277832_17278420 | 0.81 |
Tmem191c |
transmembrane protein 191C |
525 |
0.65 |
| chr15_73246575_73246818 | 0.81 |
1700085D07Rik |
RIKEN cDNA 1700085D07 gene |
2850 |
0.22 |
| chr2_68452494_68452645 | 0.80 |
Stk39 |
serine/threonine kinase 39 |
19371 |
0.19 |
| chr15_55043599_55043825 | 0.80 |
Taf2 |
TATA-box binding protein associated factor 2 |
2454 |
0.24 |
| chr1_93063621_93064051 | 0.79 |
Kif1a |
kinesin family member 1A |
4922 |
0.16 |
| chr4_64437737_64437968 | 0.79 |
Gm23950 |
predicted gene, 23950 |
6878 |
0.33 |
| chr2_104008253_104008404 | 0.79 |
Fbxo3 |
F-box protein 3 |
19393 |
0.11 |
| chr19_7196543_7197592 | 0.79 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
3397 |
0.16 |
| chr11_112873093_112873244 | 0.79 |
4933434M16Rik |
RIKEN cDNA 4933434M16 gene |
47989 |
0.15 |
| chr9_121299534_121299685 | 0.79 |
Trak1 |
trafficking protein, kinesin binding 1 |
1780 |
0.28 |
| chr5_103387516_103387737 | 0.79 |
5430427N15Rik |
RIKEN cDNA 5430427N15 gene |
30928 |
0.13 |
| chr6_108570853_108571723 | 0.78 |
Gm16433 |
predicted gene 16433 |
5700 |
0.17 |
| chr2_7519605_7519756 | 0.78 |
Celf2 |
CUGBP, Elav-like family member 2 |
10117 |
0.23 |
| chr10_81128986_81129587 | 0.78 |
Map2k2 |
mitogen-activated protein kinase kinase 2 |
671 |
0.44 |
| chr10_122870324_122870475 | 0.78 |
Ppm1h |
protein phosphatase 1H (PP2C domain containing) |
24969 |
0.19 |
| chr6_72584180_72584499 | 0.78 |
Elmod3 |
ELMO/CED-12 domain containing 3 |
10682 |
0.09 |
| chr11_35948423_35948795 | 0.77 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
31918 |
0.18 |
| chr9_91353887_91354086 | 0.77 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
3033 |
0.14 |
| chr1_65129934_65130085 | 0.77 |
D630023F18Rik |
RIKEN cDNA D630023F18 gene |
6795 |
0.12 |
| chr15_82255387_82255538 | 0.76 |
1500009C09Rik |
RIKEN cDNA 1500009C09 gene |
561 |
0.55 |
| chr15_31989572_31989923 | 0.76 |
Gm49285 |
predicted gene, 49285 |
60204 |
0.14 |
| chr4_94752503_94752932 | 0.76 |
Tek |
TEK receptor tyrosine kinase |
13199 |
0.21 |
| chr4_6840020_6840184 | 0.76 |
Tox |
thymocyte selection-associated high mobility group box |
150381 |
0.04 |
| chr6_84410254_84410424 | 0.76 |
Gm10445 |
predicted gene 10445 |
13499 |
0.26 |
| chr2_178118970_178119469 | 0.76 |
Phactr3 |
phosphatase and actin regulator 3 |
40 |
0.98 |
| chr13_72134510_72134727 | 0.76 |
Irx1 |
Iroquois homeobox 1 |
170895 |
0.03 |
| chr2_166403117_166403659 | 0.75 |
Gm14268 |
predicted gene 14268 |
5201 |
0.25 |
| chr9_34984399_34984565 | 0.75 |
Gm23702 |
predicted gene, 23702 |
24463 |
0.16 |
| chr17_81538552_81538703 | 0.75 |
Gm11096 |
predicted gene 11096 |
96864 |
0.08 |
| chr13_95443458_95443716 | 0.75 |
Crhbp |
corticotropin releasing hormone binding protein |
1244 |
0.41 |
| chr14_79204811_79205110 | 0.75 |
Gm4632 |
predicted gene 4632 |
5770 |
0.17 |
| chr2_181156387_181156681 | 0.75 |
Eef1a2 |
eukaryotic translation elongation factor 1 alpha 2 |
480 |
0.69 |
| chr3_141943276_141943438 | 0.74 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
11834 |
0.29 |
| chr17_70753475_70753647 | 0.74 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
2021 |
0.31 |
| chr14_47186589_47186864 | 0.74 |
Gch1 |
GTP cyclohydrolase 1 |
2687 |
0.15 |
| chr8_104076802_104077176 | 0.74 |
Gm8748 |
predicted gene 8748 |
5840 |
0.17 |
| chr11_63338410_63338561 | 0.74 |
Gm12286 |
predicted gene 12286 |
25174 |
0.21 |
| chr5_26991582_26992107 | 0.73 |
Gm16057 |
predicted gene 16057 |
15777 |
0.25 |
| chr10_56980420_56980571 | 0.73 |
Gm36827 |
predicted gene, 36827 |
1737 |
0.46 |
| chr13_60173703_60173877 | 0.72 |
Gm48488 |
predicted gene, 48488 |
2892 |
0.21 |
| chr8_20593052_20593376 | 0.72 |
Gm21112 |
predicted gene, 21112 |
3827 |
0.2 |
| chr2_26458386_26459048 | 0.72 |
Gm13568 |
predicted gene 13568 |
1573 |
0.21 |
| chr12_82935466_82935659 | 0.72 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
3593 |
0.28 |
| chr4_125194460_125194614 | 0.71 |
1700041M05Rik |
RIKEN cDNA 1700041M05 gene |
34060 |
0.15 |
| chr4_63742174_63742328 | 0.71 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
2862 |
0.29 |
| chr17_34549976_34550127 | 0.71 |
Btnl7-ps |
butyrophilin-like 7, pseudogene |
2400 |
0.11 |
| chrX_162363227_162363562 | 0.71 |
Gm15204 |
predicted gene 15204 |
11798 |
0.27 |
| chr3_157882081_157882268 | 0.71 |
Gm6520 |
predicted gene 6520 |
6700 |
0.15 |
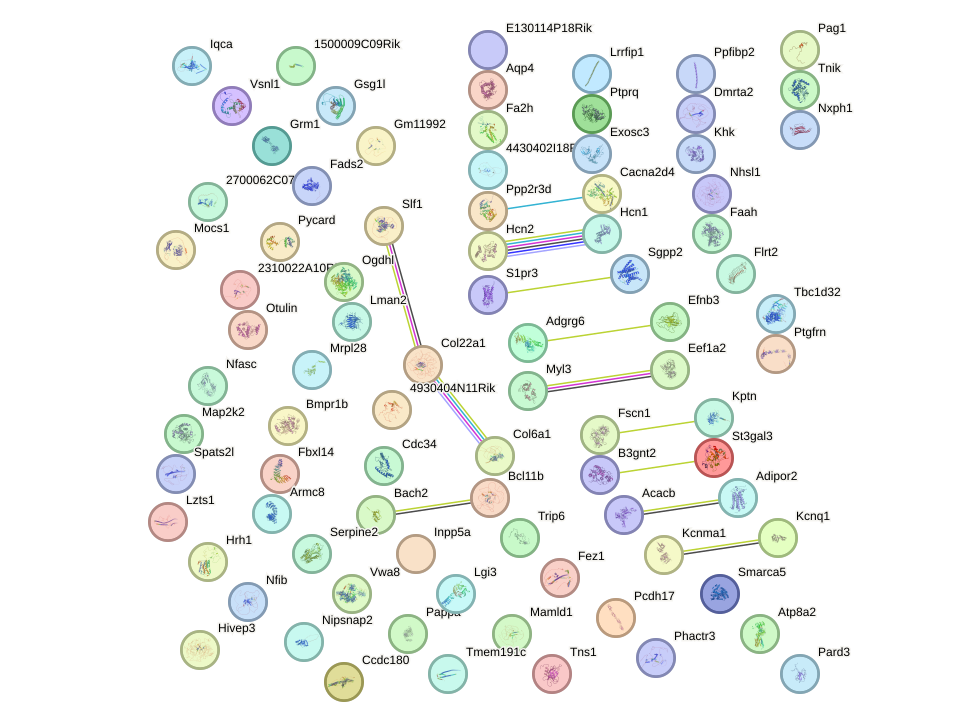
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.8 | 3.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.7 | 2.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.5 | 1.5 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.3 | 1.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.2 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.7 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.2 | 1.3 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.2 | 1.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.2 | 0.6 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 1.1 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.4 | GO:0032344 | regulation of aldosterone metabolic process(GO:0032344) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.5 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.1 | 1.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 1.0 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.3 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.4 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.4 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:1903799 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 0.5 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 1.0 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.2 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.1 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.2 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.1 | 0.4 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.6 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.5 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) ureter morphogenesis(GO:0072197) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.1 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0072173 | metanephric tubule morphogenesis(GO:0072173) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 2.4 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) regulation of eosinophil activation(GO:1902566) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:1902855 | regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.5 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0043307 | eosinophil activation(GO:0043307) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.5 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.0 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.3 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.0 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.0 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.0 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 1.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 0.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 2.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 3.8 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 1.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.2 | 0.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 0.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 3.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.2 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0018644 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.5 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) inositol-4,5-bisphosphate 5-phosphatase activity(GO:0030487) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0046030 | inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0052712 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |