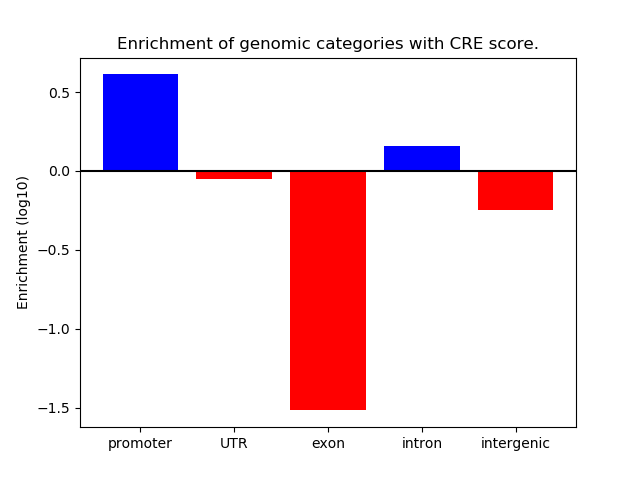
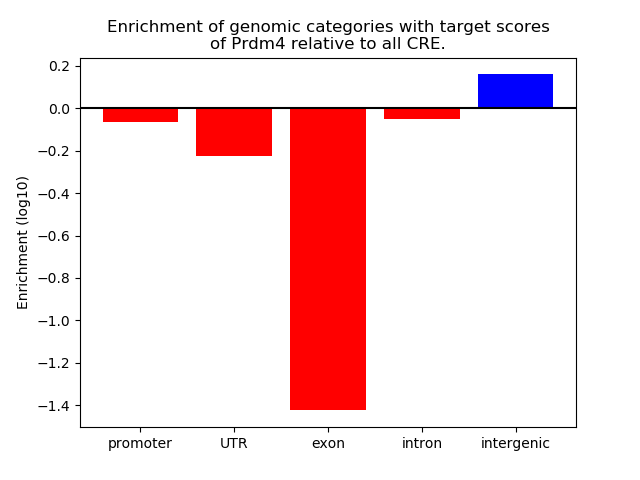
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
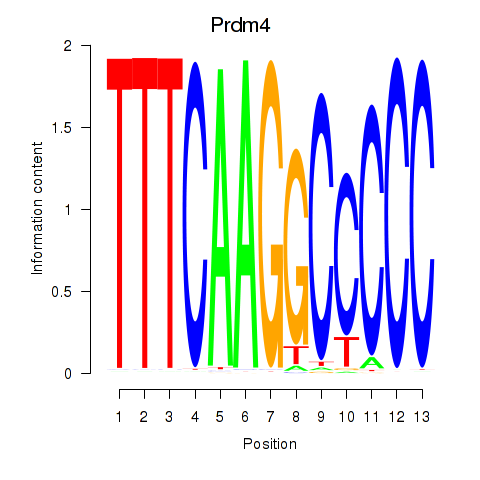
Results for Prdm4
Z-value: 0.81
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSMUSG00000035529.9 | Prdm4 |
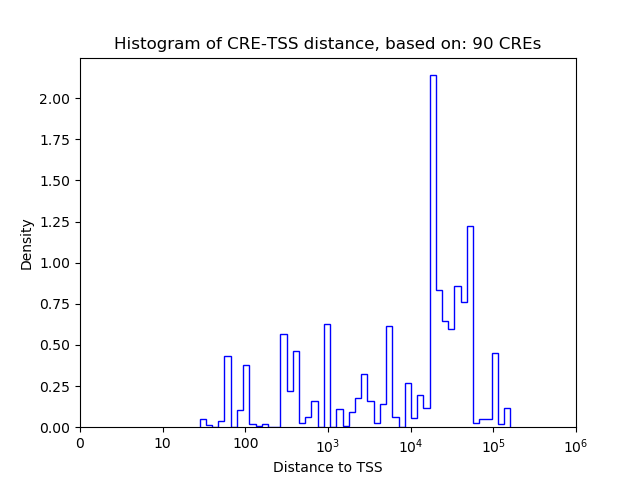
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Prdm4 | chr10_85916447_85916938 | 36 | 0.959499 | -0.03 | 8.2e-01 | Click! |
| Prdm4 | chr10_85916960_85917579 | 127 | 0.936007 | 0.03 | 8.4e-01 | Click! |
Activity of the Prdm4 motif across conditions
Conditions sorted by the z-value of the Prdm4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
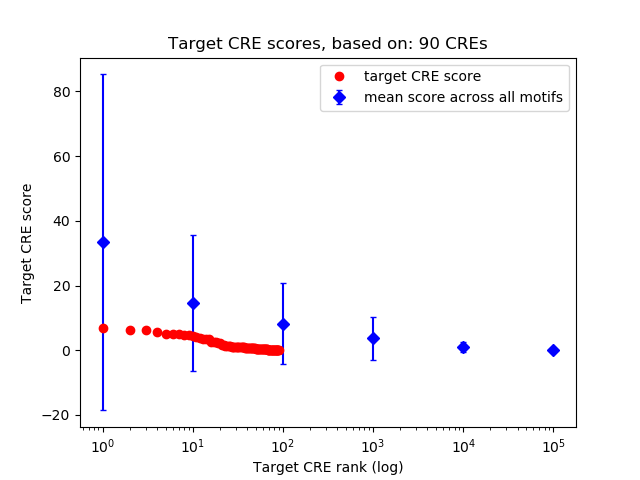
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_12716268_12716419 | 6.84 |
Gm37565 |
predicted gene, 37565 |
18926 |
0.27 |
| chr2_106895717_106896104 | 6.36 |
Gm13901 |
predicted gene 13901 |
20861 |
0.18 |
| chr19_36534720_36535517 | 6.30 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
19521 |
0.2 |
| chr10_87419886_87420133 | 5.63 |
Gm23191 |
predicted gene, 23191 |
55863 |
0.12 |
| chr14_12716331_12716619 | 5.10 |
Gm48266 |
predicted gene, 48266 |
54075 |
0.13 |
| chr2_34486214_34486378 | 5.03 |
Mapkap1 |
mitogen-activated protein kinase associated protein 1 |
41949 |
0.13 |
| chr8_83876019_83876207 | 4.95 |
Adgrl1 |
adhesion G protein-coupled receptor L1 |
23992 |
0.11 |
| chr4_98108669_98109025 | 4.76 |
Gm12691 |
predicted gene 12691 |
37752 |
0.2 |
| chr11_96872828_96873000 | 4.63 |
Gm11523 |
predicted gene 11523 |
1020 |
0.32 |
| chr10_75032772_75033005 | 4.44 |
Rsph14 |
radial spoke head homolog 14 (Chlamydomonas) |
302 |
0.83 |
| chr4_142449762_142450034 | 4.07 |
Gm13052 |
predicted gene 13052 |
103423 |
0.07 |
| chr6_87993885_87994594 | 3.90 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
60 |
0.94 |
| chr12_73476646_73476883 | 3.49 |
Gm48656 |
predicted gene, 48656 |
19857 |
0.14 |
| chr1_17145272_17145872 | 3.40 |
Gdap1 |
ganglioside-induced differentiation-associated-protein 1 |
104 |
0.97 |
| chr3_50881765_50882103 | 3.34 |
Gm37548 |
predicted gene, 37548 |
5125 |
0.23 |
| chr7_25006131_25006556 | 2.47 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
385 |
0.75 |
| chr9_77496901_77497300 | 2.42 |
Lrrc1 |
leucine rich repeat containing 1 |
35344 |
0.13 |
| chr13_78162932_78163091 | 2.40 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
8619 |
0.14 |
| chr17_86090516_86090667 | 2.32 |
Srbd1 |
S1 RNA binding domain 1 |
29680 |
0.2 |
| chr17_25415423_25415610 | 2.15 |
Cacna1h |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
17762 |
0.1 |
| chrX_166271526_166271872 | 1.74 |
Gpm6b |
glycoprotein m6b |
32727 |
0.18 |
| chr6_18446901_18447365 | 1.46 |
Gm26233 |
predicted gene, 26233 |
2207 |
0.27 |
| chr12_15803978_15804129 | 1.43 |
Mir6387 |
microRNA 6387 |
2952 |
0.22 |
| chr5_66919006_66919157 | 1.42 |
Gm43281 |
predicted gene 43281 |
3073 |
0.2 |
| chr5_100751704_100751855 | 1.36 |
Helq |
helicase, POLQ-like |
13659 |
0.12 |
| chr18_45896851_45897467 | 1.30 |
A330093E20Rik |
RIKEN cDNA A330093E20 gene |
650 |
0.79 |
| chr8_106288872_106289023 | 1.14 |
Gm10629 |
predicted gene 10629 |
5640 |
0.17 |
| chr7_45367404_45367868 | 1.12 |
Mtag2 |
metastasis associated gene 2 |
403 |
0.35 |
| chr3_104796052_104796791 | 1.08 |
Rhoc |
ras homolog family member C |
4438 |
0.11 |
| chr9_29802186_29802600 | 1.07 |
Ntm |
neurotrimin |
160726 |
0.04 |
| chr14_66911932_66912438 | 1.05 |
Pnma2 |
paraneoplastic antigen MA2 |
943 |
0.5 |
| chr19_4163611_4163806 | 1.03 |
Rps6kb2 |
ribosomal protein S6 kinase, polypeptide 2 |
354 |
0.61 |
| chr5_37857768_37858171 | 1.01 |
Msx1 |
msh homeobox 1 |
33386 |
0.15 |
| chr12_56275676_56275858 | 0.97 |
Gm47682 |
predicted gene, 47682 |
21369 |
0.14 |
| chr11_32225680_32226589 | 0.96 |
Mpg |
N-methylpurine-DNA glycosylase |
371 |
0.76 |
| chr5_135513193_135513444 | 0.91 |
Hip1 |
huntingtin interacting protein 1 |
24060 |
0.12 |
| chr15_103364800_103364960 | 0.84 |
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
1868 |
0.21 |
| chr17_43630277_43630503 | 0.76 |
Tdrd6 |
tudor domain containing 6 |
91 |
0.96 |
| chr17_16599832_16600015 | 0.74 |
Gm4786 |
predicted gene 4786 |
42677 |
0.17 |
| chr8_111033356_111033544 | 0.65 |
Aars |
alanyl-tRNA synthetase |
306 |
0.77 |
| chr12_3960175_3960559 | 0.62 |
Pomc |
pro-opiomelanocortin-alpha |
5397 |
0.16 |
| chr4_69677391_69677542 | 0.58 |
Gm11221 |
predicted gene 11221 |
14512 |
0.3 |
| chr5_92504695_92505420 | 0.57 |
Scarb2 |
scavenger receptor class B, member 2 |
580 |
0.71 |
| chr8_40962071_40962251 | 0.57 |
Pdgfrl |
platelet-derived growth factor receptor-like |
35928 |
0.12 |
| chr13_8272239_8272390 | 0.56 |
Gm48259 |
predicted gene, 48259 |
6290 |
0.16 |
| chr2_74680840_74680991 | 0.56 |
Hoxd11 |
homeobox D11 |
1358 |
0.15 |
| chr10_108589383_108589571 | 0.55 |
Syt1 |
synaptotagmin I |
47487 |
0.16 |
| chr19_56232294_56232445 | 0.54 |
Gm22271 |
predicted gene, 22271 |
45276 |
0.15 |
| chr3_69494373_69494539 | 0.53 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
2757 |
0.3 |
| chr15_36580548_36581460 | 0.49 |
Gm44310 |
predicted gene, 44310 |
2540 |
0.2 |
| chr5_20196771_20197178 | 0.48 |
Magi2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
5595 |
0.26 |
| chr15_11802561_11802718 | 0.48 |
Npr3 |
natriuretic peptide receptor 3 |
88349 |
0.07 |
| chr12_55017115_55017266 | 0.47 |
Baz1a |
bromodomain adjacent to zinc finger domain 1A |
2842 |
0.17 |
| chr14_23401177_23401361 | 0.46 |
Gm46443 |
predicted gene, 46443 |
80125 |
0.1 |
| chr3_116807637_116808371 | 0.44 |
Agl |
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
27 |
0.96 |
| chr15_30192048_30192225 | 0.43 |
Ctnnd2 |
catenin (cadherin associated protein), delta 2 |
18998 |
0.26 |
| chr14_58036330_58036481 | 0.41 |
Gm9012 |
predicted gene 9012 |
13418 |
0.16 |
| chr1_172309995_172310636 | 0.41 |
Igsf8 |
immunoglobulin superfamily, member 8 |
1456 |
0.25 |
| chr1_154190200_154190368 | 0.36 |
Zfp648 |
zinc finger protein 648 |
10903 |
0.2 |
| chr12_29654946_29655405 | 0.33 |
C630031E19Rik |
RIKEN cDNA C630031E19 gene |
31270 |
0.22 |
| chr17_90454113_90455767 | 0.33 |
Gm10493 |
predicted gene 10493 |
51 |
0.59 |
| chr7_49262055_49262574 | 0.32 |
Nav2 |
neuron navigator 2 |
15469 |
0.22 |
| chr19_48651754_48651957 | 0.31 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
52361 |
0.18 |
| chr2_39004398_39004813 | 0.29 |
Rpl35 |
ribosomal protein L35 |
438 |
0.55 |
| chr10_85101930_85102186 | 0.28 |
Fhl4 |
four and a half LIM domains 4 |
437 |
0.55 |
| chr11_12373481_12373753 | 0.24 |
Cobl |
cordon-bleu WH2 repeat |
3925 |
0.32 |
| chr18_10182315_10182716 | 0.22 |
Rock1 |
Rho-associated coiled-coil containing protein kinase 1 |
470 |
0.73 |
| chr7_119895286_119895984 | 0.18 |
Dcun1d3 |
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
92 |
0.69 |
| chr14_13519590_13519953 | 0.17 |
Synpr |
synaptoporin |
65742 |
0.13 |
| chr9_65176034_65176763 | 0.17 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
4340 |
0.15 |
| chr18_78982246_78983042 | 0.16 |
Setbp1 |
SET binding protein 1 |
126747 |
0.06 |
| chr13_63046746_63046908 | 0.16 |
Aopep |
aminopeptidase O |
21288 |
0.16 |
| chr3_89244716_89244980 | 0.15 |
Trim46 |
tripartite motif-containing 46 |
165 |
0.82 |
| chr5_147076781_147077681 | 0.15 |
Polr1d |
polymerase (RNA) I polypeptide D |
115 |
0.89 |
| chr7_4087343_4087494 | 0.15 |
Gm23741 |
predicted gene, 23741 |
14472 |
0.09 |
| chr5_71011194_71011626 | 0.14 |
Gabra2 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
2274 |
0.46 |
| chr1_161070573_161071029 | 0.13 |
Cenpl |
centromere protein L |
34 |
0.61 |
| chr1_85308637_85309019 | 0.13 |
Gm16025 |
predicted gene 16025 |
17901 |
0.11 |
| chr4_105364618_105364769 | 0.12 |
Gm12722 |
predicted gene 12722 |
10253 |
0.3 |
| chr2_90893779_90893948 | 0.12 |
Gm32514 |
predicted gene, 32514 |
665 |
0.52 |
| chr8_120290806_120290957 | 0.08 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
62425 |
0.1 |
| chr13_113589648_113589945 | 0.07 |
Gm34471 |
predicted gene, 34471 |
22029 |
0.15 |
| chr3_122072745_122072896 | 0.06 |
Abca4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
1634 |
0.37 |
| chr3_83025861_83026012 | 0.06 |
Fga |
fibrinogen alpha chain |
140 |
0.94 |
| chr10_60692133_60692313 | 0.04 |
Cdh23 |
cadherin 23 (otocadherin) |
4267 |
0.26 |
| chr18_61637596_61638529 | 0.04 |
Bvht |
braveheart long non-coding RNA |
1480 |
0.27 |
| chr7_144788527_144789274 | 0.03 |
Gm34964 |
predicted gene, 34964 |
2908 |
0.15 |
| chr6_16697126_16697277 | 0.01 |
Gm36669 |
predicted gene, 36669 |
80323 |
0.1 |
| chr1_86059453_86059994 | 0.01 |
Psmd1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
4664 |
0.13 |
| chr8_84104026_84104482 | 0.01 |
Dcaf15 |
DDB1 and CUL4 associated factor 15 |
508 |
0.56 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.5 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.2 | 3.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) activation of meiosis(GO:0090427) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.3 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.2 | GO:1903347 | apical constriction(GO:0003383) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.3 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0050861 | negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 3.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 3.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.7 | GO:0003905 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.1 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |