Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
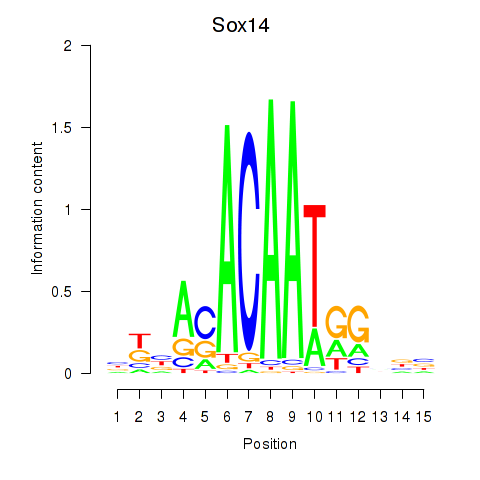
Results for Sox14
Z-value: 10.11
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSMUSG00000053747.8 | Sox14 |
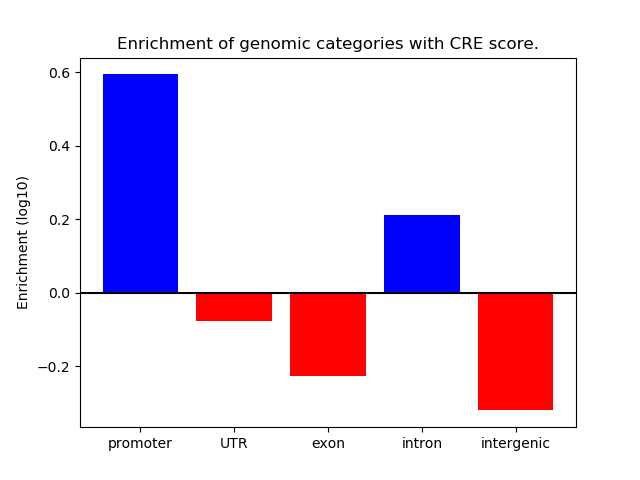
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Sox14 | chr9_99872627_99872791 | 2946 | 0.255597 | 0.85 | 1.2e-16 | Click! |
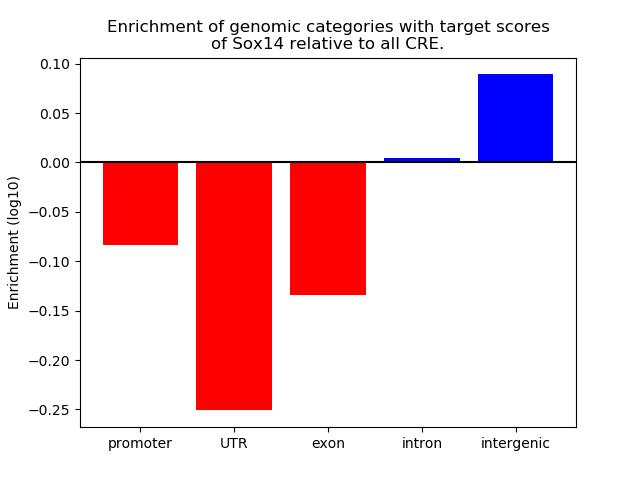
Activity of the Sox14 motif across conditions
Conditions sorted by the z-value of the Sox14 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
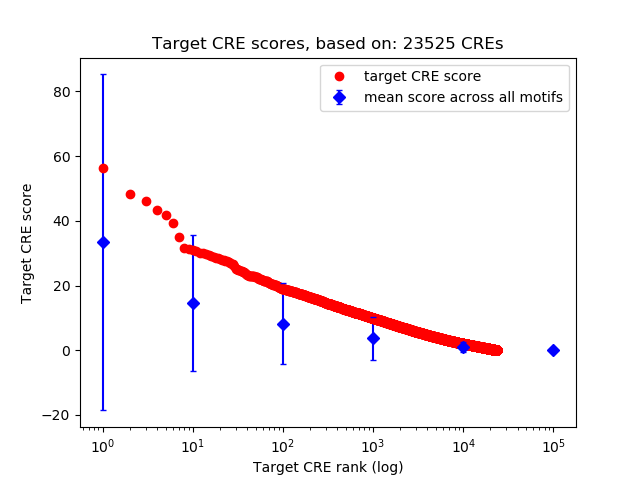
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_158062143_158062492 | 56.46 |
Lrrc40 |
leucine rich repeat containing 40 |
39 |
0.97 |
| chr16_77236189_77236366 | 48.41 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
40 |
0.98 |
| chr1_172349913_172350109 | 46.19 |
Kcnj10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
8801 |
0.11 |
| chr12_3236518_3237725 | 43.34 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
510 |
0.74 |
| chr6_89613728_89614091 | 41.79 |
Chchd6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
18257 |
0.17 |
| chr17_13590938_13591623 | 39.37 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
22416 |
0.14 |
| chr5_67752645_67752994 | 34.98 |
Gm42737 |
predicted gene 42737 |
6511 |
0.18 |
| chr18_78454680_78455309 | 31.72 |
4931439C15Rik |
RIKEN cDNA 4931439C15 gene |
21884 |
0.23 |
| chr6_40024030_40024181 | 31.23 |
Gm37995 |
predicted gene, 37995 |
2789 |
0.31 |
| chr2_50971005_50971315 | 30.97 |
Gm13498 |
predicted gene 13498 |
61476 |
0.15 |
| chr7_64533404_64533751 | 30.57 |
Gm44721 |
predicted gene 44721 |
4152 |
0.22 |
| chr13_13393240_13394314 | 30.08 |
Gpr137b |
G protein-coupled receptor 137B |
153 |
0.93 |
| chr4_3051990_3053232 | 29.95 |
Gm27878 |
predicted gene, 27878 |
792 |
0.7 |
| chrX_60544150_60544353 | 29.70 |
Gm715 |
predicted gene 715 |
3768 |
0.18 |
| chr2_105611972_105612254 | 29.40 |
Paupar |
Pax6 upstream antisense RNA |
49230 |
0.11 |
| chr10_106609605_106610022 | 29.13 |
4930532I03Rik |
RIKEN cDNA 4930532I03 gene |
7333 |
0.29 |
| chr13_12650037_12651101 | 28.81 |
Gpr137b-ps |
G protein-coupled receptor 137B, pseudogene |
181 |
0.91 |
| chr6_91345867_91346380 | 28.60 |
Wnt7a |
wingless-type MMTV integration site family, member 7A |
18948 |
0.15 |
| chr12_25871193_25871842 | 28.59 |
Gm47733 |
predicted gene, 47733 |
364 |
0.91 |
| chr16_21339491_21340087 | 28.17 |
Magef1 |
melanoma antigen family F, 1 |
6433 |
0.21 |
| chr3_159851998_159852454 | 27.91 |
Wls |
wntless WNT ligand secretion mediator |
3775 |
0.28 |
| chrX_143902215_143902612 | 27.86 |
Dcx |
doublecortin |
30637 |
0.2 |
| chr13_83741584_83742060 | 27.67 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2959 |
0.16 |
| chr18_25646197_25646553 | 27.52 |
Gm3227 |
predicted gene 3227 |
48885 |
0.16 |
| chr7_4123736_4124170 | 27.19 |
Ttyh1 |
tweety family member 1 |
536 |
0.57 |
| chr2_153425549_153426538 | 26.89 |
Gm14472 |
predicted gene 14472 |
12194 |
0.16 |
| chr9_41697271_41698297 | 26.80 |
Gm48784 |
predicted gene, 48784 |
22730 |
0.14 |
| chr2_74426718_74427115 | 26.59 |
Lnpk |
lunapark, ER junction formation factor |
106190 |
0.05 |
| chr10_3863520_3864007 | 25.94 |
Gm16149 |
predicted gene 16149 |
5844 |
0.21 |
| chr9_108825088_108825551 | 25.17 |
Gm37714 |
predicted gene, 37714 |
30 |
0.71 |
| chr8_92134225_92134404 | 24.98 |
Gm45332 |
predicted gene 45332 |
23952 |
0.17 |
| chr4_107835452_107835981 | 24.69 |
Lrp8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
4731 |
0.12 |
| chr8_53600201_53600383 | 24.65 |
Neil3 |
nei like 3 (E. coli) |
8687 |
0.27 |
| chr7_144628071_144628674 | 24.56 |
Ano1 |
anoctamin 1, calcium activated chloride channel |
8789 |
0.19 |
| chr13_39523613_39524251 | 24.35 |
Gm47351 |
predicted gene, 47351 |
9171 |
0.2 |
| chr14_76255061_76255467 | 24.20 |
2900040C04Rik |
RIKEN cDNA 2900040C04 gene |
3807 |
0.27 |
| chr18_25999104_25999542 | 24.10 |
Gm33228 |
predicted gene, 33228 |
8234 |
0.31 |
| chr19_21789708_21790094 | 23.96 |
Cemip2 |
cell migration inducing hyaluronidase 2 |
11513 |
0.21 |
| chr12_98573417_98573836 | 23.59 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
1086 |
0.43 |
| chr14_34969154_34969501 | 23.17 |
Mir346 |
microRNA 346 |
74718 |
0.11 |
| chr10_80300858_80301038 | 23.12 |
Apc2 |
APC regulator of WNT signaling pathway 2 |
86 |
0.91 |
| chr16_63806441_63806811 | 23.05 |
Epha3 |
Eph receptor A3 |
56787 |
0.15 |
| chr13_77892438_77892993 | 23.03 |
Pou5f2 |
POU domain class 5, transcription factor 2 |
132187 |
0.05 |
| chr4_23983504_23983750 | 23.02 |
Gm28448 |
predicted gene 28448 |
49673 |
0.19 |
| chr4_23787538_23787725 | 23.01 |
Gm11890 |
predicted gene 11890 |
127287 |
0.06 |
| chr13_97823915_97824432 | 22.94 |
Gm41031 |
predicted gene, 41031 |
1712 |
0.35 |
| chrX_11493886_11494235 | 22.83 |
Gm14515 |
predicted gene 14515 |
107294 |
0.06 |
| chr4_32923442_32923826 | 22.82 |
Ankrd6 |
ankyrin repeat domain 6 |
129 |
0.96 |
| chr7_109813406_109813599 | 22.59 |
Scube2 |
signal peptide, CUB domain, EGF-like 2 |
5271 |
0.15 |
| chr13_84449354_84449551 | 22.59 |
Gm26927 |
predicted gene, 26927 |
109339 |
0.06 |
| chr11_80315811_80316355 | 22.53 |
Rhbdl3 |
rhomboid like 3 |
3607 |
0.23 |
| chr3_108433472_108434154 | 22.50 |
Gm22942 |
predicted gene, 22942 |
7517 |
0.09 |
| chr9_124439906_124440949 | 22.24 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
441 |
0.79 |
| chr4_5962411_5962640 | 22.15 |
Gm11796 |
predicted gene 11796 |
107038 |
0.07 |
| chr4_122998402_122999232 | 22.10 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
63 |
0.96 |
| chr4_56128107_56128471 | 22.09 |
Gm12520 |
predicted gene 12520 |
34651 |
0.21 |
| chr3_17785697_17785901 | 21.97 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
4122 |
0.22 |
| chr12_71533009_71533369 | 21.80 |
4930404H11Rik |
RIKEN cDNA 4930404H11 gene |
7418 |
0.24 |
| chr16_90522093_90522383 | 21.77 |
Gm36001 |
predicted gene, 36001 |
28371 |
0.14 |
| chr6_10969635_10970167 | 21.74 |
AA545190 |
EST AA545190 |
4477 |
0.3 |
| chr14_103763810_103763977 | 21.50 |
Slain1os |
SLAIN motif family, member 1, opposite strand |
64451 |
0.11 |
| chr6_22834709_22835019 | 21.49 |
Gm43629 |
predicted gene 43629 |
29535 |
0.14 |
| chr7_79505823_79506244 | 21.47 |
Mir9-3 |
microRNA 9-3 |
769 |
0.43 |
| chr5_33995599_33996957 | 21.46 |
Nat8l |
N-acetyltransferase 8-like |
294 |
0.82 |
| chr13_70096512_70096709 | 21.37 |
Gm47656 |
predicted gene, 47656 |
35052 |
0.14 |
| chr14_11534595_11534746 | 21.34 |
Gm48595 |
predicted gene, 48595 |
13900 |
0.18 |
| chr18_30736874_30737155 | 21.30 |
Gm41780 |
predicted gene, 41780 |
128124 |
0.06 |
| chr2_141074597_141074768 | 21.01 |
Macrod2 |
mono-ADP ribosylhydrolase 2 |
50620 |
0.15 |
| chr11_32087496_32088333 | 20.94 |
Gm12108 |
predicted gene 12108 |
13314 |
0.2 |
| chr13_15714658_15715471 | 20.80 |
Gm48408 |
predicted gene, 48408 |
55056 |
0.11 |
| chr3_70483210_70483372 | 20.75 |
Gm6631 |
predicted gene 6631 |
69972 |
0.12 |
| chr16_91962945_91963627 | 20.74 |
Gm27773 |
predicted gene, 27773 |
19138 |
0.1 |
| chr6_112929919_112930359 | 20.48 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
16615 |
0.14 |
| chr1_51617502_51617987 | 20.48 |
Gm17767 |
predicted gene, 17767 |
21026 |
0.21 |
| chr15_78770837_78771346 | 20.45 |
Mfng |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
803 |
0.52 |
| chr3_13470653_13471071 | 20.41 |
Ralyl |
RALY RNA binding protein-like |
793 |
0.48 |
| chr12_12287368_12287790 | 20.41 |
Fam49a |
family with sequence similarity 49, member A |
25390 |
0.24 |
| chrX_36195676_36196141 | 20.31 |
Zcchc12 |
zinc finger, CCHC domain containing 12 |
4 |
0.98 |
| chr11_111766566_111766769 | 20.23 |
Gm11674 |
predicted gene 11674 |
34209 |
0.2 |
| chr3_4546148_4546485 | 20.23 |
Gm22944 |
predicted gene, 22944 |
21019 |
0.21 |
| chr18_28078106_28078315 | 20.17 |
Gm5064 |
predicted gene 5064 |
110607 |
0.07 |
| chr4_72233972_72234136 | 20.15 |
C630043F03Rik |
RIKEN cDNA C630043F03 gene |
32706 |
0.18 |
| chr16_64069361_64069662 | 20.07 |
Gm49627 |
predicted gene, 49627 |
114381 |
0.07 |
| chr11_32157683_32158170 | 20.05 |
Gm12109 |
predicted gene 12109 |
27079 |
0.12 |
| chr10_17330771_17331243 | 19.95 |
Gm47760 |
predicted gene, 47760 |
1870 |
0.39 |
| chr9_36821980_36822332 | 19.87 |
Fez1 |
fasciculation and elongation protein zeta 1 (zygin I) |
292 |
0.87 |
| chr8_123411336_123412017 | 19.85 |
Tubb3 |
tubulin, beta 3 class III |
86 |
0.91 |
| chr12_98014890_98015065 | 19.77 |
Gm35412 |
predicted gene, 35412 |
85378 |
0.08 |
| chr3_157321061_157321436 | 19.61 |
Gm22458 |
predicted gene, 22458 |
44750 |
0.14 |
| chr3_126994207_126994654 | 19.57 |
Ank2 |
ankyrin 2, brain |
4008 |
0.14 |
| chr12_50120364_50120537 | 19.41 |
Gm40418 |
predicted gene, 40418 |
141 |
0.98 |
| chr4_54652498_54652694 | 19.40 |
Gm12480 |
predicted gene 12480 |
693 |
0.64 |
| chr16_33605736_33606716 | 19.23 |
Slc12a8 |
solute carrier family 12 (potassium/chloride transporters), member 8 |
9599 |
0.26 |
| chr1_6733683_6734408 | 19.17 |
St18 |
suppression of tumorigenicity 18 |
825 |
0.73 |
| chr10_81059624_81060601 | 19.11 |
Sgta |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
3 |
0.94 |
| chr14_24617943_24618207 | 19.09 |
4930428N03Rik |
RIKEN cDNA 4930428N03 gene |
507 |
0.52 |
| chr3_83098548_83099077 | 19.03 |
Dchs2 |
dachsous cadherin related 2 |
29136 |
0.13 |
| chr6_134888153_134888812 | 19.00 |
Gpr19 |
G protein-coupled receptor 19 |
650 |
0.56 |
| chr17_88764144_88764434 | 18.99 |
Lhcgr |
luteinizing hormone/choriogonadotropin receptor |
9767 |
0.24 |
| chr15_36141846_36142049 | 18.96 |
Rgs22 |
regulator of G-protein signalling 22 |
1547 |
0.27 |
| chr2_69418258_69418920 | 18.92 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
38144 |
0.15 |
| chr13_15543962_15544319 | 18.90 |
Gli3 |
GLI-Kruppel family member GLI3 |
80160 |
0.08 |
| chr7_91208818_91209022 | 18.87 |
Gm24552 |
predicted gene, 24552 |
34075 |
0.14 |
| chr4_96697628_96698496 | 18.83 |
Cyp2j5 |
cytochrome P450, family 2, subfamily j, polypeptide 5 |
33908 |
0.19 |
| chr2_67488575_67488726 | 18.80 |
Gm13601 |
predicted gene 13601 |
27875 |
0.15 |
| chr11_112811583_112811789 | 18.78 |
Gm11681 |
predicted gene 11681 |
12678 |
0.18 |
| chr9_102243320_102243774 | 18.76 |
Gm37260 |
predicted gene, 37260 |
30189 |
0.16 |
| chr8_64153329_64153548 | 18.73 |
Gm22586 |
predicted gene, 22586 |
7967 |
0.26 |
| chr12_12694102_12694394 | 18.72 |
Gm25538 |
predicted gene, 25538 |
5791 |
0.17 |
| chr6_55681352_55681586 | 18.71 |
Neurod6 |
neurogenic differentiation 6 |
206 |
0.95 |
| chr14_100374770_100375216 | 18.66 |
Gm26367 |
predicted gene, 26367 |
43490 |
0.15 |
| chr3_94478560_94479074 | 18.65 |
Celf3 |
CUGBP, Elav-like family member 3 |
14 |
0.94 |
| chr9_94838531_94838931 | 18.65 |
Mir7656 |
microRNA 7656 |
86902 |
0.09 |
| chr1_88245559_88246456 | 18.62 |
Mroh2a |
maestro heat-like repeat family member 2A |
7899 |
0.09 |
| chr13_36612886_36613098 | 18.54 |
Gm46409 |
predicted gene, 46409 |
20 |
0.97 |
| chr15_24567016_24567356 | 18.48 |
Gm48920 |
predicted gene, 48920 |
38292 |
0.19 |
| chr18_73096635_73097020 | 18.43 |
Gm31908 |
predicted gene, 31908 |
175737 |
0.03 |
| chr8_47288315_47288537 | 18.42 |
Stox2 |
storkhead box 2 |
936 |
0.65 |
| chr9_45663374_45663610 | 18.41 |
Dscaml1 |
DS cell adhesion molecule like 1 |
9345 |
0.19 |
| chr4_22835787_22836371 | 18.40 |
Gm24078 |
predicted gene, 24078 |
88948 |
0.09 |
| chr9_13652356_13652901 | 18.37 |
Maml2 |
mastermind like transcriptional coactivator 2 |
9832 |
0.2 |
| chr15_34824497_34824736 | 18.35 |
Gm48932 |
predicted gene, 48932 |
1813 |
0.39 |
| chr10_18470489_18470661 | 18.34 |
Nhsl1 |
NHS-like 1 |
594 |
0.81 |
| chr2_157340114_157340758 | 18.33 |
Ghrh |
growth hormone releasing hormone |
3010 |
0.22 |
| chr15_7194818_7195373 | 18.29 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
27974 |
0.21 |
| chr18_64014262_64014492 | 18.27 |
Gm6974 |
predicted gene 6974 |
67043 |
0.1 |
| chr8_35933319_35934071 | 18.24 |
Gm22030 |
predicted gene, 22030 |
43883 |
0.13 |
| chr9_41918970_41919408 | 18.14 |
Gm40513 |
predicted gene, 40513 |
28585 |
0.14 |
| chr19_46523220_46524148 | 18.09 |
Trim8 |
tripartite motif-containing 8 |
9024 |
0.15 |
| chr12_12578319_12578604 | 18.07 |
AC107703.1 |
novel transcript |
91456 |
0.07 |
| chr2_113828734_113829247 | 18.00 |
Scg5 |
secretogranin V |
131 |
0.96 |
| chr6_50776731_50777167 | 17.98 |
C530044C16Rik |
RIKEN cDNA C530044C16 gene |
625 |
0.69 |
| chr5_112643537_112643881 | 17.98 |
4933415J04Rik |
RIKEN cDNA 4933415J04 gene |
3374 |
0.19 |
| chr8_94567491_94567722 | 17.97 |
Cpne2 |
copine II |
4956 |
0.14 |
| chr7_96718538_96719200 | 17.95 |
Tenm4 |
teneurin transmembrane protein 4 |
59065 |
0.11 |
| chr3_17789882_17790060 | 17.95 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
14 |
0.98 |
| chr2_4036647_4036830 | 17.92 |
Gm2639 |
predicted gene 2639 |
15821 |
0.14 |
| chr16_45723055_45723347 | 17.92 |
Tagln3 |
transgelin 3 |
1407 |
0.33 |
| chr9_72924923_72926161 | 17.86 |
Pygo1 |
pygopus 1 |
103 |
0.93 |
| chr4_105171033_105171295 | 17.81 |
Plpp3 |
phospholipid phosphatase 3 |
13817 |
0.26 |
| chr14_79880455_79880704 | 17.78 |
Gm6999 |
predicted gene 6999 |
10418 |
0.16 |
| chr2_152080641_152081210 | 17.78 |
Scrt2 |
scratch family zinc finger 2 |
604 |
0.66 |
| chr6_66399103_66399403 | 17.78 |
Gm44233 |
predicted gene, 44233 |
507 |
0.47 |
| chr4_109225194_109225586 | 17.76 |
Calr4 |
calreticulin 4 |
9095 |
0.2 |
| chr7_73184982_73185667 | 17.68 |
Gm28746 |
predicted gene 28746 |
69 |
0.86 |
| chr16_63091512_63091690 | 17.68 |
Gm49621 |
predicted gene, 49621 |
20089 |
0.25 |
| chr17_52601056_52601789 | 17.65 |
Gm27217 |
predicted gene 27217 |
1238 |
0.39 |
| chr16_35581258_35581604 | 17.60 |
Gm5963 |
predicted pseudogene 5963 |
7875 |
0.19 |
| chr10_57828634_57829003 | 17.52 |
Smpdl3a |
sphingomyelin phosphodiesterase, acid-like 3A |
34250 |
0.16 |
| chr16_89530496_89531006 | 17.49 |
Krtap7-1 |
keratin associated protein 7-1 |
22428 |
0.15 |
| chr5_38158989_38159808 | 17.46 |
Nsg1 |
neuron specific gene family member 1 |
7 |
0.97 |
| chr1_5019058_5019249 | 17.46 |
Rgs20 |
regulator of G-protein signaling 20 |
226 |
0.92 |
| chr12_55537902_55538053 | 17.41 |
Aldoart2 |
aldolase 1 A, retrogene 2 |
27262 |
0.14 |
| chr13_84448589_84449243 | 17.41 |
Gm26927 |
predicted gene, 26927 |
108803 |
0.07 |
| chr5_103335878_103336033 | 17.40 |
Gm42619 |
predicted gene 42619 |
8017 |
0.16 |
| chr10_13117108_13117586 | 17.39 |
Plagl1 |
pleiomorphic adenoma gene-like 1 |
983 |
0.59 |
| chr9_90693038_90693253 | 17.38 |
Gm2497 |
predicted gene 2497 |
40379 |
0.14 |
| chr3_35353984_35354177 | 17.37 |
Gm25442 |
predicted gene, 25442 |
28466 |
0.2 |
| chr10_87502418_87502577 | 17.33 |
Gm48120 |
predicted gene, 48120 |
5365 |
0.2 |
| chr9_63182441_63182600 | 17.32 |
Skor1 |
SKI family transcriptional corepressor 1 |
33559 |
0.15 |
| chr2_6875430_6875726 | 17.26 |
Celf2 |
CUGBP, Elav-like family member 2 |
2981 |
0.26 |
| chr14_118416177_118416529 | 17.24 |
Gm5672 |
predicted gene 5672 |
40159 |
0.12 |
| chr7_60170912_60171242 | 17.22 |
Gm7367 |
predicted pseudogene 7367 |
15468 |
0.13 |
| chr13_20473180_20473431 | 17.18 |
Gm32036 |
predicted gene, 32036 |
185 |
0.82 |
| chr9_96980387_96981016 | 17.16 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
3494 |
0.21 |
| chr8_36155388_36156182 | 17.13 |
Gm38414 |
predicted gene, 38414 |
225 |
0.92 |
| chr13_107540300_107540691 | 17.11 |
Gm32004 |
predicted gene, 32004 |
24828 |
0.2 |
| chr11_32002527_32003111 | 17.10 |
Nsg2 |
neuron specific gene family member 2 |
2317 |
0.33 |
| chr10_29145120_29145386 | 17.08 |
Gm9996 |
predicted gene 9996 |
1059 |
0.37 |
| chr19_26978313_26978606 | 17.07 |
Gm50121 |
predicted gene, 50121 |
47522 |
0.16 |
| chr2_45225367_45225747 | 17.01 |
Gm28643 |
predicted gene 28643 |
68632 |
0.11 |
| chr2_70126801_70127190 | 16.99 |
Myo3b |
myosin IIIB |
30697 |
0.2 |
| chr16_79176820_79177259 | 16.97 |
Tmprss15 |
transmembrane protease, serine 15 |
85942 |
0.1 |
| chr2_57267160_57267354 | 16.96 |
Gpd2 |
glycerol phosphate dehydrogenase 2, mitochondrial |
120 |
0.96 |
| chr3_35090595_35090820 | 16.94 |
Mir6378 |
microRNA 6378 |
168056 |
0.03 |
| chr5_37245988_37246869 | 16.94 |
Crmp1 |
collapsin response mediator protein 1 |
583 |
0.76 |
| chr4_55294236_55294477 | 16.93 |
Gm25419 |
predicted gene, 25419 |
5489 |
0.16 |
| chr8_30648481_30648881 | 16.88 |
4933433F19Rik |
RIKEN cDNA 4933433F19 gene |
80288 |
0.11 |
| chr1_118913010_118913408 | 16.87 |
Mir6346 |
microRNA 6346 |
41011 |
0.16 |
| chr18_26753599_26753876 | 16.81 |
Gm49976 |
predicted gene, 49976 |
164360 |
0.04 |
| chr13_59092007_59092498 | 16.78 |
4930415C11Rik |
RIKEN cDNA 4930415C11 gene |
8159 |
0.17 |
| chr1_6757654_6757828 | 16.77 |
St18 |
suppression of tumorigenicity 18 |
20166 |
0.24 |
| chr12_51002470_51002650 | 16.73 |
Gm40421 |
predicted gene, 40421 |
2313 |
0.29 |
| chr10_81230482_81230696 | 16.67 |
Atcay |
ataxia, cerebellar, Cayman type |
196 |
0.83 |
| chr4_25408022_25408173 | 16.67 |
Gm11894 |
predicted gene 11894 |
11252 |
0.22 |
| chr15_67245652_67245820 | 16.65 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
18967 |
0.26 |
| chr15_66239660_66240017 | 16.65 |
Kcnq3 |
potassium voltage-gated channel, subfamily Q, member 3 |
46213 |
0.14 |
| chr8_17587417_17587764 | 16.64 |
Csmd1 |
CUB and Sushi multiple domains 1 |
52004 |
0.19 |
| chr17_15380918_15381780 | 16.59 |
Dll1 |
delta like canonical Notch ligand 1 |
4477 |
0.18 |
| chr11_112869991_112870994 | 16.58 |
4933434M16Rik |
RIKEN cDNA 4933434M16 gene |
45313 |
0.16 |
| chr3_68513686_68514094 | 16.57 |
Schip1 |
schwannomin interacting protein 1 |
19682 |
0.2 |
| chr13_83714747_83715651 | 16.55 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
6182 |
0.14 |
| chr10_119684387_119684717 | 16.55 |
Grip1 |
glutamate receptor interacting protein 1 |
7502 |
0.21 |
| chrX_49273592_49273893 | 16.51 |
Enox2 |
ecto-NOX disulfide-thiol exchanger 2 |
14470 |
0.24 |
| chr16_90565374_90565525 | 16.49 |
1110008E08Rik |
RIKEN cDNA 1110008E08 gene |
11285 |
0.15 |
| chr13_48171687_48171898 | 16.46 |
Gm36346 |
predicted gene, 36346 |
65459 |
0.09 |
| chr1_57083188_57083527 | 16.45 |
9130024F11Rik |
RIKEN cDNA 9130024F11 gene |
42964 |
0.14 |
| chr5_130924338_130924528 | 16.40 |
Gm42897 |
predicted gene 42897 |
25820 |
0.18 |
| chr4_53317418_53317807 | 16.38 |
Gm12495 |
predicted gene 12495 |
9386 |
0.21 |
| chr2_151701768_151703133 | 16.33 |
Tmem74b |
transmembrane protein 74B |
139 |
0.92 |
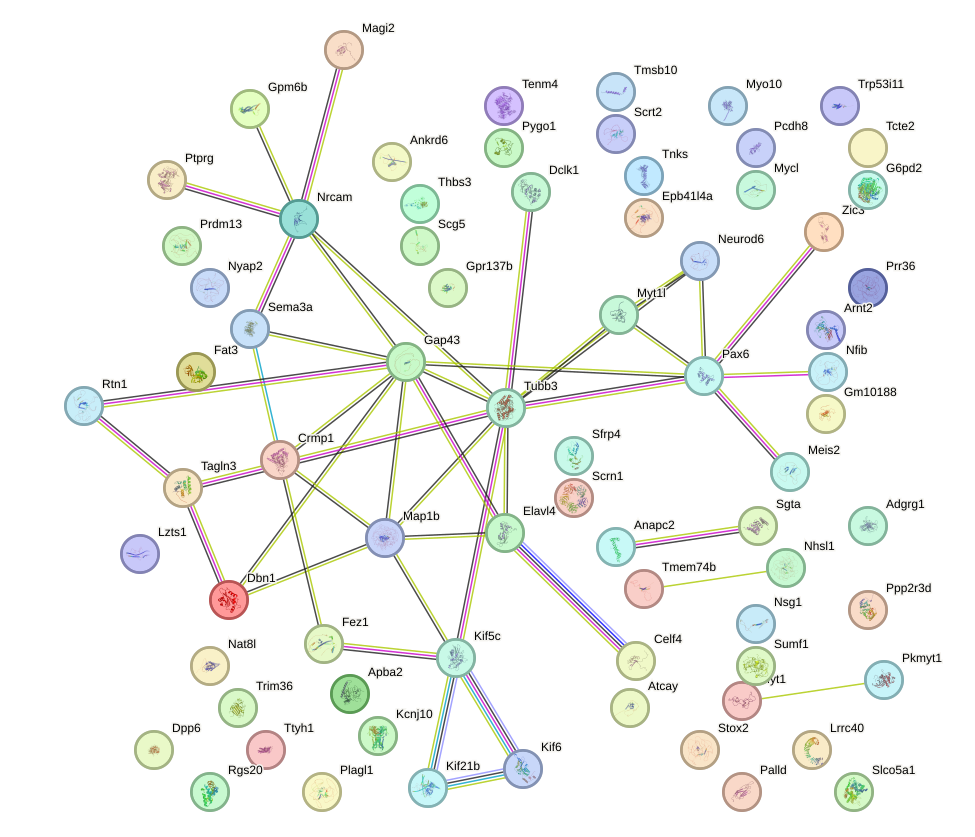
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.1 | 75.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 10.8 | 10.8 | GO:2000981 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 10.4 | 31.2 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 10.3 | 30.8 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 8.7 | 34.9 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 7.4 | 22.3 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 7.3 | 22.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 7.1 | 21.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 7.0 | 21.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 6.6 | 13.3 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 6.5 | 45.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 6.4 | 25.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 6.1 | 24.4 | GO:0060594 | mammary gland specification(GO:0060594) |
| 6.1 | 24.2 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 6.0 | 18.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 6.0 | 18.0 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 5.9 | 17.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 5.9 | 23.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 5.8 | 17.3 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 5.6 | 16.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 5.5 | 21.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 5.5 | 16.4 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 5.4 | 27.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 5.4 | 43.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 5.4 | 21.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 5.3 | 26.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 5.2 | 20.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 5.2 | 15.5 | GO:0097503 | sialylation(GO:0097503) |
| 5.1 | 51.3 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 5.1 | 15.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 4.9 | 14.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 4.8 | 19.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 4.8 | 14.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 4.5 | 36.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 4.4 | 4.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 4.4 | 17.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 4.4 | 17.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 4.3 | 21.7 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 4.3 | 51.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 4.3 | 12.9 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 4.2 | 16.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 4.2 | 29.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 4.2 | 8.3 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 4.1 | 45.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 4.1 | 12.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 4.1 | 8.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 3.7 | 7.4 | GO:0048880 | sensory system development(GO:0048880) |
| 3.7 | 21.9 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 3.6 | 10.9 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 3.6 | 3.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 3.6 | 14.5 | GO:0060174 | limb bud formation(GO:0060174) |
| 3.6 | 7.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 3.5 | 17.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 3.5 | 70.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 3.5 | 13.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 3.5 | 10.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 3.4 | 10.3 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 3.4 | 6.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 3.4 | 16.9 | GO:0060278 | regulation of ovulation(GO:0060278) |
| 3.4 | 20.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 3.4 | 6.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 3.4 | 10.1 | GO:0030070 | insulin processing(GO:0030070) |
| 3.3 | 10.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 3.3 | 10.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 3.3 | 13.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 3.3 | 9.9 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 3.2 | 16.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 3.2 | 12.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 3.2 | 9.6 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 3.2 | 6.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 3.2 | 9.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 3.1 | 9.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 3.1 | 12.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 3.1 | 3.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 3.1 | 12.4 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 3.1 | 39.8 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 3.0 | 6.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 3.0 | 15.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 3.0 | 9.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 3.0 | 15.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 3.0 | 12.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 3.0 | 8.9 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 3.0 | 3.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 3.0 | 8.9 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 2.9 | 5.9 | GO:0060166 | olfactory pit development(GO:0060166) |
| 2.9 | 32.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 2.9 | 23.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 2.9 | 23.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 2.8 | 8.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 2.8 | 17.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 2.8 | 8.5 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 2.8 | 8.5 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 2.8 | 8.4 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 2.8 | 2.8 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 2.7 | 8.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 2.7 | 13.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 2.7 | 13.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.7 | 16.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 2.6 | 10.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.6 | 5.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 2.6 | 7.9 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 2.6 | 13.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 2.6 | 5.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 2.6 | 10.2 | GO:0046959 | habituation(GO:0046959) |
| 2.6 | 2.6 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 2.6 | 10.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 2.5 | 2.5 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 2.5 | 43.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 2.5 | 5.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 2.5 | 19.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 2.4 | 14.6 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 2.4 | 9.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 2.4 | 7.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 2.4 | 4.8 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 2.4 | 7.2 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 2.4 | 4.8 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 2.4 | 4.8 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 2.4 | 7.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 2.4 | 9.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 2.3 | 7.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.3 | 4.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.3 | 9.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 2.3 | 2.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 2.2 | 6.7 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 2.2 | 8.9 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 2.2 | 11.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 2.2 | 4.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.2 | 2.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 2.2 | 19.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 2.2 | 19.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 2.2 | 17.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 2.1 | 23.6 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 2.1 | 6.4 | GO:0060437 | lung growth(GO:0060437) |
| 2.1 | 30.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 2.1 | 6.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 2.1 | 33.5 | GO:0060384 | innervation(GO:0060384) |
| 2.1 | 8.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 2.1 | 22.8 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 2.1 | 2.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 2.1 | 8.2 | GO:0030091 | protein repair(GO:0030091) |
| 2.1 | 16.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 2.0 | 12.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.0 | 2.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 2.0 | 2.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 2.0 | 8.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 2.0 | 10.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 2.0 | 13.8 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 2.0 | 2.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 2.0 | 17.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 2.0 | 19.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 2.0 | 2.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.9 | 1.9 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 1.9 | 5.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 1.9 | 5.7 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.9 | 5.7 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.9 | 3.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.9 | 7.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.9 | 1.9 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 1.9 | 7.5 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 1.9 | 5.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.9 | 3.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 1.8 | 5.5 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 1.8 | 3.7 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 1.8 | 14.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.8 | 9.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 1.8 | 5.4 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 1.8 | 3.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.8 | 5.3 | GO:0061356 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 1.8 | 17.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 1.8 | 3.5 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 1.8 | 7.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 1.8 | 5.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 1.8 | 17.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 1.7 | 1.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.7 | 5.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 1.7 | 5.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.7 | 3.4 | GO:0072174 | metanephric tubule formation(GO:0072174) |
| 1.7 | 1.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.7 | 28.4 | GO:0001964 | startle response(GO:0001964) |
| 1.7 | 16.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.6 | 3.3 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.6 | 4.9 | GO:0072017 | distal tubule development(GO:0072017) |
| 1.6 | 11.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 1.6 | 3.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.6 | 1.6 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 1.6 | 6.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 1.6 | 9.5 | GO:0060581 | ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 1.6 | 3.2 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 1.6 | 7.8 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 1.6 | 3.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 1.5 | 1.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 1.5 | 3.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 1.5 | 4.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 1.5 | 4.6 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.5 | 3.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.5 | 3.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.5 | 4.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 1.5 | 4.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.5 | 10.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.5 | 1.5 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.5 | 12.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.5 | 7.5 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 1.5 | 1.5 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.5 | 4.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.5 | 4.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 1.5 | 2.9 | GO:0051665 | membrane raft localization(GO:0051665) |
| 1.5 | 4.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.5 | 4.4 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 1.4 | 8.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 1.4 | 4.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 1.4 | 1.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 1.4 | 11.5 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 1.4 | 2.8 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 1.4 | 22.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.4 | 1.4 | GO:0048382 | mesendoderm development(GO:0048382) |
| 1.4 | 1.4 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.4 | 4.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 1.4 | 107.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 1.4 | 2.8 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 1.4 | 31.7 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 1.4 | 4.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.4 | 4.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 1.3 | 4.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 1.3 | 4.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 1.3 | 10.7 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 1.3 | 2.7 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.3 | 2.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 1.3 | 46.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 1.3 | 5.2 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 1.3 | 6.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 1.3 | 3.8 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 1.3 | 2.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.3 | 6.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.3 | 3.8 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.3 | 1.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 1.2 | 3.7 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.2 | 2.5 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 1.2 | 3.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.2 | 2.5 | GO:0071435 | potassium ion export(GO:0071435) |
| 1.2 | 3.7 | GO:0061743 | motor learning(GO:0061743) |
| 1.2 | 2.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 1.2 | 3.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 1.2 | 4.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.2 | 4.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.2 | 2.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 1.2 | 10.9 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 1.2 | 2.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.2 | 7.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.2 | 4.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 1.2 | 3.6 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.2 | 2.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 1.2 | 14.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 1.2 | 2.3 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 1.2 | 3.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 1.2 | 3.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 1.2 | 2.3 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 1.2 | 2.3 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 1.2 | 1.2 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.2 | 2.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 1.1 | 4.6 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 1.1 | 1.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.1 | 4.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.1 | 3.4 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 1.1 | 4.5 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 1.1 | 4.5 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 1.1 | 17.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 1.1 | 3.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.1 | 3.3 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 1.1 | 3.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 1.1 | 26.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 1.1 | 2.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.1 | 2.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.1 | 2.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.1 | 6.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 1.1 | 5.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 1.1 | 2.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.1 | 6.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 1.1 | 4.2 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 1.0 | 1.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.0 | 17.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 1.0 | 7.2 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 1.0 | 31.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 1.0 | 5.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 1.0 | 7.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.0 | 3.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.0 | 12.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 1.0 | 4.1 | GO:0060013 | righting reflex(GO:0060013) |
| 1.0 | 1.0 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 1.0 | 3.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.0 | 6.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.0 | 3.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.0 | 9.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 1.0 | 14.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.0 | 1.0 | GO:0098815 | modulation of excitatory postsynaptic potential(GO:0098815) |
| 1.0 | 2.0 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 1.0 | 2.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.0 | 7.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 1.0 | 2.9 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.0 | 1.9 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 1.0 | 15.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.0 | 13.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 1.0 | 3.9 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 1.0 | 1.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 1.0 | 2.9 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 1.0 | 1.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.0 | 4.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 1.0 | 2.9 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.9 | 4.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.9 | 1.9 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.9 | 14.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.9 | 11.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.9 | 4.6 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.9 | 4.6 | GO:0032196 | transposition(GO:0032196) |
| 0.9 | 1.8 | GO:0030432 | peristalsis(GO:0030432) |
| 0.9 | 2.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.9 | 1.8 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.9 | 3.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.9 | 4.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.9 | 3.5 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.9 | 3.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.9 | 22.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.9 | 3.5 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.9 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.9 | 3.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.9 | 3.4 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.9 | 8.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.9 | 1.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.9 | 0.9 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.8 | 5.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.8 | 1.7 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.8 | 2.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.8 | 2.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.8 | 1.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.8 | 2.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.8 | 7.3 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.8 | 1.6 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.8 | 7.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.8 | 0.8 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.8 | 2.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.8 | 4.8 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.8 | 2.4 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.8 | 1.6 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.8 | 1.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.8 | 10.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.8 | 3.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.8 | 4.7 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.8 | 0.8 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.8 | 6.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.8 | 2.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.8 | 24.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.8 | 0.8 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.8 | 0.8 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.8 | 2.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.8 | 1.5 | GO:2000412 | thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.8 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.8 | 2.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.8 | 1.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.7 | 2.2 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.7 | 2.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 | 6.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.7 | 4.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.7 | 7.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.7 | 2.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.7 | 2.9 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.7 | 9.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.7 | 3.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.7 | 2.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.7 | 1.4 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.7 | 2.1 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.7 | 2.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.7 | 2.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.7 | 4.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.7 | 2.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.7 | 1.4 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.7 | 1.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.7 | 2.1 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.7 | 2.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.7 | 2.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.7 | 4.7 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.7 | 3.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.7 | 1.3 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.7 | 12.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.7 | 0.7 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.7 | 4.0 | GO:0060004 | reflex(GO:0060004) |
| 0.7 | 0.7 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.7 | 2.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.7 | 2.0 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.7 | 2.0 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.7 | 5.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.7 | 1.3 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.6 | 0.6 | GO:0021586 | pons maturation(GO:0021586) |
| 0.6 | 3.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.6 | 18.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 0.6 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.6 | 3.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.6 | 2.5 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.6 | 1.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 3.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.6 | 3.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.6 | 0.6 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.6 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.6 | 3.7 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.6 | 1.8 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.6 | 1.8 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.6 | 1.2 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.6 | 1.2 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.6 | 1.2 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.6 | 3.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.6 | 4.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.6 | 1.2 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) skeletal muscle satellite cell activation(GO:0014719) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.6 | 0.6 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.6 | 1.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 1.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 17.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 0.6 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.6 | 0.6 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.6 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.6 | 1.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.6 | 1.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.6 | 2.8 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.6 | 5.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.6 | 6.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.5 | 1.6 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.5 | 1.6 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.5 | 0.5 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 2.7 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.5 | 2.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.5 | 7.5 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.5 | 1.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.5 | 1.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 2.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.5 | 0.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.5 | 5.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.5 | 1.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.5 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 3.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.5 | 1.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.5 | 0.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.5 | 1.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 4.6 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.5 | 1.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.5 | 2.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.5 | 1.5 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.5 | 3.0 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.5 | 2.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.5 | 1.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.5 | 1.0 | GO:0001975 | response to amphetamine(GO:0001975) |
| 0.5 | 1.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 3.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 3.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.5 | 1.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.5 | 1.0 | GO:0070268 | cornification(GO:0070268) |
| 0.5 | 24.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.5 | 1.9 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 11.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.5 | 28.1 | GO:0007612 | learning(GO:0007612) |
| 0.5 | 5.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.5 | 0.9 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.5 | 10.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.5 | 1.8 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 1.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 1.4 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.4 | 0.9 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.4 | 3.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 1.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.4 | 1.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 1.7 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.4 | 0.9 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 | 0.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.4 | 2.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 1.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.4 | 2.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 1.3 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.4 | 0.4 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.4 | 3.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 2.5 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.4 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.4 | 0.8 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.4 | 0.4 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.4 | 2.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 1.2 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.4 | 4.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 1.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.4 | 2.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 7.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.4 | 0.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 0.8 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.4 | 3.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.4 | 2.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.4 | 0.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 0.4 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.4 | 4.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 0.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.4 | 1.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 2.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 0.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.4 | 1.2 | GO:0019042 | viral latency(GO:0019042) |
| 0.4 | 1.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.4 | 1.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 0.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.4 | 0.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 0.8 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.4 | 2.6 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.4 | 0.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.4 | 0.7 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.4 | 1.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 2.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.4 | 1.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.4 | 1.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.3 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 1.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 0.3 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.3 | 1.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.3 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.3 | 0.7 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 3.0 | GO:0036093 | germ cell proliferation(GO:0036093) |
| 0.3 | 0.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 1.3 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.3 | 0.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.3 | 2.6 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 4.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.3 | 1.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 1.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 0.6 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.3 | 1.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 0.9 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.3 | 0.6 | GO:0032725 | granulocyte macrophage colony-stimulating factor production(GO:0032604) regulation of granulocyte macrophage colony-stimulating factor production(GO:0032645) positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.3 | 0.6 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 3.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.3 | 0.9 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.3 | 1.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 1.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 0.3 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.3 | 0.3 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 0.6 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.3 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 2.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 1.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 0.9 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.3 | 3.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 0.9 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.3 | 0.9 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.3 | 3.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 | 1.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 0.9 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.3 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 1.1 | GO:0034982 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.3 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.3 | 1.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 2.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 1.4 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.3 | 0.6 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.3 | 0.8 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 | 0.8 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 3.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 0.8 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 0.8 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.3 | 1.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 1.0 | GO:0001963 | synaptic transmission, dopaminergic(GO:0001963) |
| 0.3 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.3 | 0.5 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.3 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.3 | 0.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 1.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 1.4 | GO:0043576 | regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 1.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.2 | 0.5 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.2 | 2.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 3.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.2 | GO:0071611 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.2 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.2 | 0.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 1.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.7 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 2.4 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.2 | 0.4 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 | 0.2 | GO:0032532 | parallel actin filament bundle assembly(GO:0030046) regulation of microvillus length(GO:0032532) |
| 0.2 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 1.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.6 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.2 | 1.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 7.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 0.6 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 1.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.4 | GO:0015810 | aspartate transport(GO:0015810) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.6 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 2.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.4 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.2 | 0.5 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.2 | 0.5 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.4 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.2 | 0.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.2 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 0.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 0.3 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 0.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 6.6 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.3 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.2 | 1.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 0.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.2 | 0.3 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.2 | 0.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.2 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.2 | 3.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 0.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.4 | GO:0014841 | skeletal muscle satellite cell proliferation(GO:0014841) |
| 0.1 | 2.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.3 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.1 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.0 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.1 | 0.8 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.5 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 0.1 | GO:0021783 | preganglionic parasympathetic fiber development(GO:0021783) |
| 0.1 | 2.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.1 | 0.5 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.3 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.1 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.1 | 0.5 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.1 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.1 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.2 | GO:0060452 | positive regulation of cardiac muscle contraction(GO:0060452) |
| 0.1 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 0.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.1 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.1 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.1 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.7 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.9 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 3.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.0 | 0.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 24.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 6.5 | 32.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 5.8 | 17.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 5.3 | 36.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 5.2 | 15.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 5.2 | 15.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 5.1 | 55.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 4.8 | 19.2 | GO:0044308 | axonal spine(GO:0044308) |
| 4.8 | 9.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 4.5 | 35.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 4.3 | 12.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 4.1 | 24.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 4.0 | 28.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 3.8 | 15.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 3.7 | 29.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 3.5 | 28.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 2.7 | 23.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 2.6 | 44.8 | GO:0030673 | axolemma(GO:0030673) |
| 2.5 | 30.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 2.5 | 7.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 2.5 | 29.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 2.4 | 9.6 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 2.4 | 45.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 2.3 | 2.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 2.2 | 15.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 2.2 | 55.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.2 | 6.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 2.1 | 19.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 2.1 | 12.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 2.1 | 85.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 2.0 | 12.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 2.0 | 7.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.9 | 7.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.9 | 7.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 1.8 | 14.7 | GO:0071437 | invadopodium(GO:0071437) |
| 1.7 | 5.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.7 | 3.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.7 | 29.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.7 | 12.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.7 | 6.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.6 | 1.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.6 | 22.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.6 | 4.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 1.5 | 4.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.5 | 4.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 1.4 | 4.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.4 | 33.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.4 | 4.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.4 | 6.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.3 | 14.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.3 | 6.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.3 | 5.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.3 | 6.4 | GO:0071547 | piP-body(GO:0071547) |
| 1.3 | 6.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.3 | 2.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.3 | 6.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 5.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.2 | 32.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 1.2 | 3.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.2 | 4.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.1 | 5.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.1 | 3.4 | GO:0043511 | inhibin complex(GO:0043511) |
| 1.1 | 4.5 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.1 | 10.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.1 | 9.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 169.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 1.1 | 3.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.1 | 6.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.1 | 6.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 1.0 | 3.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.0 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 8.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.0 | 1.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.0 | 3.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 1.0 | 10.7 | GO:0042555 | MCM complex(GO:0042555) |
| 1.0 | 4.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 16.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.9 | 6.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.9 | 1.8 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.9 | 2.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.9 | 36.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.8 | 77.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.8 | 2.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.7 | 1.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.7 | 2.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 1.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.7 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.7 | 2.7 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.7 | 1.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 3.9 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.7 | 1.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.6 | 5.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 3.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.6 | 2.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.6 | 1.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.6 | 2.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.6 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 8.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 5.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.6 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.5 | 1.6 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.5 | 1.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.5 | 1.6 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.5 | 31.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.5 | 3.6 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 1.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.5 | 17.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.5 | 25.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.5 | 3.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 1.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 1.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.5 | 6.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 1.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 3.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.4 | 34.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.4 | 1.3 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 3.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.4 | 1.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 1.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.4 | 4.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.4 | 1.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 1.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 2.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 2.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.4 | 1.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 1.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 1.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 14.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 1.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.4 | 48.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 3.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 8.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 8.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 2.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 18.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.3 | 2.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.3 | 2.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 3.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 1.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 7.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 0.8 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.3 | 2.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 2.4 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.2 | 2.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 1.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.8 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 1.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 11.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 21.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 2.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.1 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.4 | 49.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 12.1 | 36.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 9.9 | 29.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 9.3 | 37.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 6.5 | 19.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 6.2 | 30.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 5.8 | 28.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 5.4 | 16.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 5.2 | 25.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 5.2 | 15.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 5.1 | 35.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 4.6 | 18.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 4.6 | 18.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 4.4 | 17.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 4.3 | 12.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 4.1 | 12.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 4.0 | 7.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 3.9 | 15.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 3.8 | 11.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 3.8 | 11.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 3.8 | 11.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 3.7 | 22.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 3.6 | 14.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 3.5 | 24.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 3.5 | 20.9 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 3.4 | 10.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 3.2 | 16.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 3.1 | 15.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 3.0 | 15.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 3.0 | 11.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 2.9 | 8.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.9 | 17.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 2.8 | 5.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 2.7 | 8.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 2.7 | 16.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 2.7 | 10.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 2.5 | 33.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 2.4 | 7.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 2.4 | 47.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 2.4 | 9.4 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 2.3 | 27.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 2.2 | 6.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.1 | 4.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 2.1 | 46.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 2.0 | 20.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 2.0 | 22.1 | GO:0005522 | profilin binding(GO:0005522) |
| 2.0 | 27.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 2.0 | 11.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.9 | 9.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.8 | 7.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.8 | 5.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.7 | 5.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.7 | 10.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.7 | 5.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.7 | 1.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 1.7 | 15.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 1.7 | 3.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.7 | 3.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 1.7 | 6.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.6 | 6.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.6 | 38.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 1.6 | 6.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 1.5 | 19.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.5 | 30.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.5 | 9.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 1.5 | 4.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 1.5 | 10.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.5 | 19.4 | GO:0031005 | filamin binding(GO:0031005) |
| 1.5 | 4.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.5 | 4.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 1.5 | 54.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 1.5 | 5.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 1.4 | 15.9 | GO:0070694 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 1.4 | 5.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.4 | 2.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.4 | 8.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.3 | 4.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.3 | 9.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 1.3 | 14.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.3 | 14.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.3 | 10.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.3 | 7.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.3 | 5.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.2 | 6.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.2 | 3.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.2 | 3.6 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 1.2 | 5.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.2 | 13.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 1.2 | 3.5 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.2 | 3.5 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 1.2 | 29.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 1.2 | 18.6 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 1.1 | 16.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 1.1 | 10.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 1.1 | 10.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.1 | 5.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.1 | 38.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 1.1 | 5.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 1.1 | 3.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.1 | 16.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 1.1 | 3.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 1.1 | 10.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 1.1 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.1 | 24.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.1 | 17.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 1.1 | 3.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.1 | 3.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 1.1 | 4.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 1.0 | 20.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.0 | 10.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.0 | 3.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.0 | 5.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 1.0 | 9.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 1.0 | 6.0 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 1.0 | 3.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.0 | 2.9 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.0 | 1.9 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 1.0 | 1.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.0 | 4.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.9 | 3.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.9 | 1.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.9 | 1.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.9 | 0.9 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.9 | 5.7 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.9 | 22.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.9 | 16.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.9 | 0.9 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.9 | 9.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.9 | 19.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.9 | 8.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.9 | 7.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 10.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.9 | 12.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.9 | 0.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.9 | 4.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.8 | 7.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.8 | 2.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.8 | 3.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.8 | 1.7 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.8 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.8 | 10.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.8 | 15.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.8 | 3.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 4.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 6.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.8 | 2.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.8 | 6.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.8 | 2.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.8 | 2.3 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.8 | 5.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.8 | 5.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.8 | 2.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.8 | 3.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.7 | 3.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.7 | 6.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 4.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.7 | 7.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.7 | 15.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.7 | 2.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 1.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.7 | 0.7 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.7 | 2.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.7 | 3.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 2.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.7 | 6.9 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.7 | 1.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.7 | 6.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.7 | 13.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.7 | 2.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.6 | 3.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.6 | 0.6 | GO:0003905 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.6 | 13.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 3.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.6 | 1.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.6 | 3.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.6 | 4.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.6 | 1.8 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.6 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.6 | 9.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.6 | 1.8 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.6 | 2.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.6 | 2.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.6 | 8.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.6 | 1.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.6 | 13.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 5.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.6 | 8.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 3.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 2.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.5 | 9.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.5 | 2.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 2.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 15.3 | GO:0052770 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.5 | 2.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 5.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.5 | 7.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.5 | 4.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 2.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 1.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.5 | 2.8 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.5 | 4.5 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.4 | 1.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 1.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 8.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.4 | 1.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 1.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.4 | 9.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.4 | 3.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 3.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.4 | 5.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 2.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 0.8 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.4 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.4 | 9.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 0.4 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.4 | 6.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 2.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 3.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 1.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 43.7 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.4 | 3.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 15.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.4 | 1.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 10.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.4 | 4.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 0.7 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.4 | 1.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.4 | 1.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 1.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.4 | 1.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 1.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 1.4 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 7.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 11.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 0.7 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 0.6 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 5.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 2.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.3 | 6.3 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.3 | 0.9 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.3 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 0.8 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 5.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 0.5 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.2 | 13.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 1.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.7 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 3.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 3.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.2 | 2.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 1.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 3.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.2 | 0.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 5.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 0.9 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.2 | 7.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 0.6 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 1.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 4.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.7 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 0.9 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 0.5 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.2 | 0.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 5.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.1 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 2.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.7 | GO:0016937 | short-branched-chain-acyl-CoA dehydrogenase activity(GO:0016937) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.2 | GO:0046428 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 14.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 14.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 36.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 2.0 | 58.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 1.5 | 48.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 1.4 | 1.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 1.1 | 3.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 1.1 | 14.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 1.0 | 12.5 | ST ADRENERGIC | Adrenergic Pathway |
| 1.0 | 11.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.0 | 23.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.9 | 13.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 19.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.8 | 13.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.7 | 13.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.7 | 19.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.7 | 17.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.6 | 14.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.5 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.5 | 8.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.5 | 18.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.4 | 8.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 10.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 12.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 1.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 14.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 5.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 3.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 12.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 6.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 9.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 9.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 95.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 3.2 | 6.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 2.7 | 43.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 2.5 | 25.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 2.4 | 2.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 2.4 | 30.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 2.3 | 33.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 2.2 | 2.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 2.2 | 23.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 2.1 | 2.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 2.1 | 4.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 2.0 | 26.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 2.0 | 34.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 2.0 | 14.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 2.0 | 2.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 1.9 | 26.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.8 | 34.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 1.8 | 21.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.8 | 7.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.7 | 20.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.6 | 69.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 1.5 | 4.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.5 | 33.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 1.5 | 33.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.3 | 27.5 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 1.3 | 10.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 1.3 | 8.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 1.2 | 13.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.2 | 4.6 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 1.1 | 35.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 1.1 | 4.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.1 | 9.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 1.1 | 11.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.1 | 14.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 1.0 | 2.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.0 | 1.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 1.0 | 16.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 1.0 | 7.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 18.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.9 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.8 | 16.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.8 | 10.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.8 | 5.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.8 | 10.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.8 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.7 | 10.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.7 | 4.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.6 | 14.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.6 | 15.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.6 | 4.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.6 | 9.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.6 | 12.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 6.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.5 | 11.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.5 | 5.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 8.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 8.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 15.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.5 | 0.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.5 | 5.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 2.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.5 | 9.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 2.2 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.4 | 8.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 1.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.4 | 2.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.4 | 3.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 1.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 5.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 3.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 2.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.2 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 1.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 0.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 4.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.2 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.9 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |