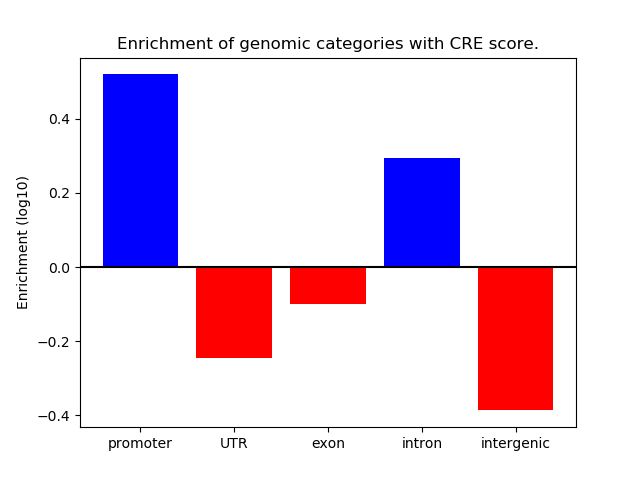
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
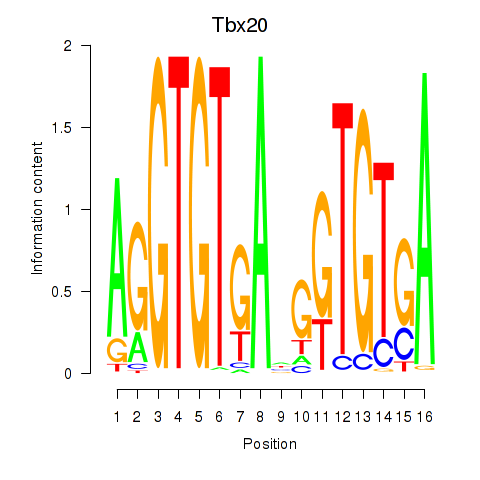
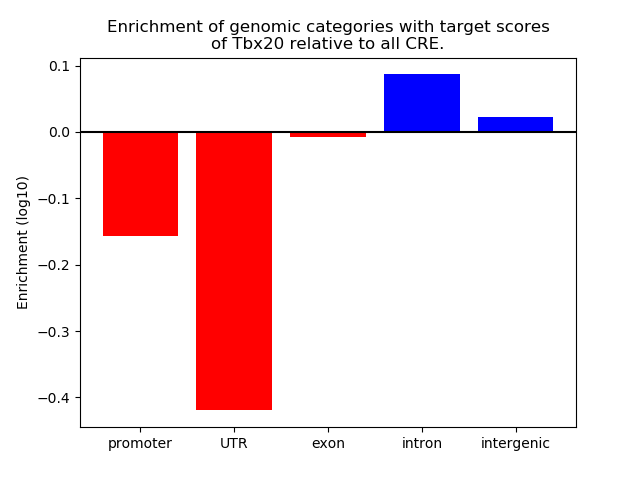
Results for Tbx20
Z-value: 1.26
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSMUSG00000031965.8 | Tbx20 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Tbx20 | chr9_24774667_24774833 | 447 | 0.779968 | 0.39 | 3.4e-03 | Click! |
| Tbx20 | chr9_24773768_24774563 | 119 | 0.958780 | 0.33 | 1.4e-02 | Click! |
| Tbx20 | chr9_24774976_24775185 | 777 | 0.546620 | 0.31 | 2.1e-02 | Click! |
| Tbx20 | chr9_24770408_24770624 | 766 | 0.647308 | 0.21 | 1.3e-01 | Click! |
Activity of the Tbx20 motif across conditions
Conditions sorted by the z-value of the Tbx20 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_13654565_13655321 | 28.70 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
13948 |
0.15 |
| chr14_6637139_6637428 | 5.70 |
Gm3460 |
predicted gene 3460 |
14865 |
0.14 |
| chr17_13590288_13590503 | 5.00 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
21531 |
0.15 |
| chr13_46918635_46918792 | 4.88 |
Kif13a |
kinesin family member 13A |
11004 |
0.13 |
| chr17_13590938_13591623 | 4.75 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
22416 |
0.14 |
| chr19_38054215_38055320 | 4.71 |
I830134H01Rik |
RIKEN cDNA I830134H01 gene |
239 |
0.48 |
| chr10_122025202_122025630 | 4.66 |
Srgap1 |
SLIT-ROBO Rho GTPase activating protein 1 |
21892 |
0.15 |
| chr9_58847804_58848021 | 4.36 |
Hcn4 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 4 |
24500 |
0.2 |
| chr2_48380305_48380493 | 3.78 |
Gm25338 |
predicted gene, 25338 |
3939 |
0.26 |
| chrX_167012030_167012225 | 3.76 |
Gm22368 |
predicted gene, 22368 |
16870 |
0.16 |
| chr17_3299883_3300126 | 3.46 |
Gm6475 |
predicted gene 6475 |
14181 |
0.15 |
| chr5_113249673_113249824 | 3.36 |
Tmem211 |
transmembrane protein 211 |
22839 |
0.12 |
| chr10_5289382_5289831 | 3.34 |
Gm23573 |
predicted gene, 23573 |
67567 |
0.12 |
| chr6_59261862_59262174 | 3.29 |
Tigd2 |
tigger transposable element derived 2 |
53148 |
0.14 |
| chrX_160588887_160589038 | 3.28 |
Phka2 |
phosphorylase kinase alpha 2 |
31175 |
0.17 |
| chr5_34217838_34218173 | 3.08 |
Gm42848 |
predicted gene 42848 |
3157 |
0.14 |
| chr13_92758294_92758445 | 3.02 |
Thbs4 |
thrombospondin 4 |
16605 |
0.19 |
| chr1_57302006_57302319 | 2.99 |
Gm5254 |
predicted gene 5254 |
9761 |
0.21 |
| chr12_76046714_76046865 | 2.98 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
1789 |
0.44 |
| chr14_4969050_4969254 | 2.96 |
Gm8297 |
predicted gene 8297 |
14783 |
0.12 |
| chr7_112660416_112660763 | 2.96 |
Gm45473 |
predicted gene 45473 |
7807 |
0.16 |
| chr1_39721483_39721634 | 2.89 |
Rfx8 |
regulatory factor X 8 |
561 |
0.78 |
| chr9_115356751_115357118 | 2.87 |
Mir467h |
microRNA 467h |
24885 |
0.11 |
| chr8_103234369_103234717 | 2.83 |
Gm32531 |
predicted gene, 32531 |
36848 |
0.2 |
| chr8_13621025_13621176 | 2.79 |
Rasa3 |
RAS p21 protein activator 3 |
14992 |
0.18 |
| chr7_130982381_130982744 | 2.77 |
Htra1 |
HtrA serine peptidase 1 |
168 |
0.96 |
| chr14_121494258_121494578 | 2.74 |
Slc15a1 |
solute carrier family 15 (oligopeptide transporter), member 1 |
1949 |
0.3 |
| chr14_19610871_19611070 | 2.73 |
4930469B13Rik |
RIKEN cDNA 4930469B13 gene |
4160 |
0.12 |
| chr1_43247602_43248038 | 2.71 |
Gm29610 |
predicted gene 29610 |
22863 |
0.16 |
| chr1_99069837_99069988 | 2.66 |
Gm23434 |
predicted gene, 23434 |
52548 |
0.18 |
| chr5_114508052_114508203 | 2.65 |
Gm13790 |
predicted gene 13790 |
43162 |
0.1 |
| chr10_117987810_117988005 | 2.65 |
Gm37852 |
predicted gene, 37852 |
17031 |
0.19 |
| chr12_79534856_79535007 | 2.64 |
Rad51b |
RAD51 paralog B |
207578 |
0.02 |
| chr12_73408705_73408856 | 2.57 |
D830013O20Rik |
RIKEN cDNA D830013O20 gene |
777 |
0.62 |
| chr13_15602594_15602999 | 2.47 |
Gm48343 |
predicted gene, 48343 |
47139 |
0.16 |
| chr9_44498071_44499796 | 2.41 |
Bcl9l |
B cell CLL/lymphoma 9-like |
203 |
0.84 |
| chr17_29237992_29238373 | 2.35 |
Cpne5 |
copine V |
15 |
0.96 |
| chr16_24487648_24487799 | 2.31 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
39632 |
0.15 |
| chr18_11044554_11044882 | 2.27 |
Gata6os |
GATA binding protein 6, opposite strand |
2919 |
0.29 |
| chr4_146484532_146484780 | 2.26 |
Gm13245 |
predicted gene 13245 |
3989 |
0.16 |
| chr13_44232185_44232493 | 2.26 |
Gm47781 |
predicted gene, 47781 |
59 |
0.97 |
| chr9_41936734_41936932 | 2.22 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
37754 |
0.13 |
| chr17_83109790_83110198 | 2.21 |
Gm46606 |
predicted gene, 46606 |
5641 |
0.22 |
| chr10_25982771_25982945 | 2.18 |
Tmem200a |
transmembrane protein 200A |
34685 |
0.16 |
| chr14_3545899_3546050 | 2.14 |
Gm3005 |
predicted gene 3005 |
26049 |
0.11 |
| chr15_85724908_85725104 | 2.13 |
Ppara |
peroxisome proliferator activated receptor alpha |
9977 |
0.13 |
| chr14_19610521_19610738 | 2.12 |
4930469B13Rik |
RIKEN cDNA 4930469B13 gene |
4501 |
0.11 |
| chr11_45831770_45831921 | 2.11 |
Gm22751 |
predicted gene, 22751 |
2066 |
0.22 |
| chr10_81006336_81006487 | 2.06 |
Gng7 |
guanine nucleotide binding protein (G protein), gamma 7 |
5033 |
0.1 |
| chr16_35439200_35439360 | 2.01 |
Gm25548 |
predicted gene, 25548 |
1083 |
0.46 |
| chr15_76816855_76818442 | 1.99 |
Arhgap39 |
Rho GTPase activating protein 39 |
322 |
0.82 |
| chr17_87611074_87611259 | 1.97 |
Epcam |
epithelial cell adhesion molecule |
24813 |
0.15 |
| chr2_159053534_159053847 | 1.97 |
Gm44319 |
predicted gene, 44319 |
16173 |
0.26 |
| chr13_8597762_8597921 | 1.94 |
Gm48262 |
predicted gene, 48262 |
44821 |
0.17 |
| chr2_163361015_163361186 | 1.93 |
Jph2 |
junctophilin 2 |
36849 |
0.11 |
| chr5_24973304_24973523 | 1.92 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
12429 |
0.18 |
| chr11_4302766_4302953 | 1.90 |
Gm11958 |
predicted gene 11958 |
17399 |
0.12 |
| chr11_86807230_86808078 | 1.88 |
Dhx40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
92 |
0.98 |
| chr17_83588253_83588542 | 1.86 |
Kcng3 |
potassium voltage-gated channel, subfamily G, member 3 |
43498 |
0.16 |
| chr7_105787328_105788039 | 1.85 |
Dchs1 |
dachsous cadherin related 1 |
29 |
0.95 |
| chr2_180391070_180391221 | 1.84 |
Mir1a-1 |
microRNA 1a-1 |
2097 |
0.22 |
| chr5_33495974_33496125 | 1.84 |
Gm43851 |
predicted gene 43851 |
58585 |
0.09 |
| chr3_142918167_142918318 | 1.84 |
Gm31306 |
predicted gene, 31306 |
20167 |
0.13 |
| chrX_94049185_94049336 | 1.84 |
Gm14798 |
predicted gene 14798 |
20406 |
0.14 |
| chr9_123844308_123844459 | 1.81 |
Fyco1 |
FYVE and coiled-coil domain containing 1 |
223 |
0.9 |
| chr17_12045961_12046332 | 1.80 |
Prkn |
parkin RBR E3 ubiquitin protein ligase |
16230 |
0.24 |
| chr13_114478670_114478821 | 1.80 |
Fst |
follistatin |
19794 |
0.15 |
| chr7_129664007_129664606 | 1.80 |
Gm33248 |
predicted gene, 33248 |
3099 |
0.26 |
| chr2_140113926_140114077 | 1.80 |
Gm14073 |
predicted gene 14073 |
3599 |
0.21 |
| chr8_8639759_8639910 | 1.79 |
Efnb2 |
ephrin B2 |
471 |
0.48 |
| chr3_50185958_50186323 | 1.79 |
Gm37692 |
predicted gene, 37692 |
19248 |
0.24 |
| chr11_98348139_98348301 | 1.72 |
Ppp1r1b |
protein phosphatase 1, regulatory inhibitor subunit 1B |
184 |
0.88 |
| chr12_12334579_12334761 | 1.71 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
57945 |
0.14 |
| chr3_73390133_73390284 | 1.69 |
Gm38353 |
predicted gene, 38353 |
60774 |
0.14 |
| chr1_120082880_120083497 | 1.66 |
Tmem37 |
transmembrane protein 37 |
9114 |
0.18 |
| chr15_58472722_58472886 | 1.62 |
Fam91a1 |
family with sequence similarity 91, member A1 |
30071 |
0.16 |
| chr17_66502983_66503134 | 1.62 |
Rab12 |
RAB12, member RAS oncogene family |
2739 |
0.24 |
| chr14_23243627_23243778 | 1.61 |
Gm46443 |
predicted gene, 46443 |
77442 |
0.12 |
| chr17_47276763_47276914 | 1.60 |
Gm26216 |
predicted gene, 26216 |
14118 |
0.17 |
| chr15_57761631_57761782 | 1.60 |
9330154K18Rik |
RIKEN cDNA 9330154K18 gene |
23040 |
0.19 |
| chr3_132945799_132945950 | 1.58 |
Npnt |
nephronectin |
3940 |
0.19 |
| chr5_35621167_35621318 | 1.58 |
Acox3 |
acyl-Coenzyme A oxidase 3, pristanoyl |
11143 |
0.15 |
| chr10_125961290_125962183 | 1.57 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
4432 |
0.33 |
| chr4_108838012_108838203 | 1.56 |
Txndc12 |
thioredoxin domain containing 12 (endoplasmic reticulum) |
3472 |
0.17 |
| chr9_118319094_118319336 | 1.56 |
2610509F24Rik |
RIKEN cDNA 2610509F24 gene |
84087 |
0.08 |
| chr6_125529706_125529887 | 1.55 |
Vwf |
Von Willebrand factor |
16978 |
0.15 |
| chr1_39410534_39410685 | 1.55 |
Gm37265 |
predicted gene, 37265 |
9323 |
0.16 |
| chr16_44558403_44558958 | 1.53 |
Boc |
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
217 |
0.79 |
| chr9_107554133_107555386 | 1.51 |
Rassf1 |
Ras association (RalGDS/AF-6) domain family member 1 |
176 |
0.81 |
| chr18_9464563_9464714 | 1.51 |
Ccny |
cyclin Y |
14484 |
0.15 |
| chr14_77621987_77622138 | 1.50 |
Gm49003 |
predicted gene, 49003 |
11798 |
0.21 |
| chr12_44401957_44402486 | 1.50 |
Gm48182 |
predicted gene, 48182 |
7569 |
0.2 |
| chr4_106688599_106689103 | 1.45 |
Ttc4 |
tetratricopeptide repeat domain 4 |
9907 |
0.13 |
| chr12_84884811_84884980 | 1.45 |
Gm48709 |
predicted gene, 48709 |
3071 |
0.16 |
| chr11_35283014_35283508 | 1.44 |
Gm12123 |
predicted gene 12123 |
40409 |
0.17 |
| chr5_53107124_53107388 | 1.43 |
n-R5s171 |
nuclear encoded rRNA 5S 171 |
35904 |
0.13 |
| chr10_110921940_110922160 | 1.41 |
Csrp2 |
cysteine and glycine-rich protein 2 |
1845 |
0.28 |
| chr12_95695278_95696175 | 1.40 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
369 |
0.86 |
| chr13_98684440_98684591 | 1.40 |
Tmem171 |
transmembrane protein 171 |
10253 |
0.14 |
| chr4_36183692_36183983 | 1.40 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
47374 |
0.2 |
| chr5_120656418_120656764 | 1.38 |
Rasal1 |
RAS protein activator like 1 (GAP1 like) |
3224 |
0.12 |
| chr4_45011783_45012858 | 1.38 |
Zbtb5 |
zinc finger and BTB domain containing 5 |
74 |
0.87 |
| chr6_34599917_34600068 | 1.37 |
Cald1 |
caldesmon 1 |
1372 |
0.45 |
| chr10_4599076_4599227 | 1.36 |
Esr1 |
estrogen receptor 1 (alpha) |
12442 |
0.2 |
| chr2_172906256_172906593 | 1.36 |
Bmp7 |
bone morphogenetic protein 7 |
33668 |
0.15 |
| chr9_76035167_76035347 | 1.35 |
Gm26903 |
predicted gene, 26903 |
20065 |
0.15 |
| chr9_58084691_58085027 | 1.33 |
Ccdc33 |
coiled-coil domain containing 33 |
2790 |
0.19 |
| chr17_55477478_55477674 | 1.33 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
31332 |
0.19 |
| chr11_113052633_113053064 | 1.32 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
120229 |
0.06 |
| chr4_119991034_119991187 | 1.30 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
41656 |
0.17 |
| chr8_30643165_30643406 | 1.29 |
4933433F19Rik |
RIKEN cDNA 4933433F19 gene |
74892 |
0.12 |
| chr4_13364740_13365135 | 1.29 |
Gm11819 |
predicted gene 11819 |
79833 |
0.1 |
| chr2_38460869_38461233 | 1.28 |
Gm25081 |
predicted gene, 25081 |
19287 |
0.12 |
| chr5_17573775_17573992 | 1.27 |
Sema3c |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
398 |
0.9 |
| chr2_164754933_164755088 | 1.26 |
Dnttip1 |
deoxynucleotidyltransferase, terminal, interacting protein 1 |
871 |
0.34 |
| chr1_164461825_164462045 | 1.25 |
Gm32391 |
predicted gene, 32391 |
507 |
0.72 |
| chr10_81416800_81417494 | 1.24 |
Mir1191b |
microRNA 1191b |
850 |
0.3 |
| chr18_61666034_61666747 | 1.24 |
Carmn |
cardiac mesoderm enhancer-associated non-coding RNA |
853 |
0.44 |
| chr2_90496145_90496296 | 1.23 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
16970 |
0.17 |
| chr3_151551098_151551520 | 1.23 |
Adgrl4 |
adhesion G protein-coupled receptor L4 |
33880 |
0.19 |
| chr9_58739924_58740075 | 1.22 |
Rec114 |
REC114 meiotic recombination protein |
1560 |
0.44 |
| chr1_135800050_135801209 | 1.21 |
Tnni1 |
troponin I, skeletal, slow 1 |
796 |
0.55 |
| chr8_85554792_85555798 | 1.20 |
Dnaja2 |
DnaJ heat shock protein family (Hsp40) member A2 |
49 |
0.97 |
| chr9_32674301_32674452 | 1.20 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
21646 |
0.15 |
| chr18_8044886_8045210 | 1.19 |
Gm4833 |
predicted gene 4833 |
6155 |
0.28 |
| chr6_103944408_103944702 | 1.18 |
Gm21054 |
predicted gene, 21054 |
20084 |
0.27 |
| chr19_15827405_15827866 | 1.17 |
Gm50348 |
predicted gene, 50348 |
24577 |
0.22 |
| chr10_90722912_90723063 | 1.17 |
Gm47591 |
predicted gene, 47591 |
5973 |
0.2 |
| chr11_86457684_86457835 | 1.17 |
Gm26439 |
predicted gene, 26439 |
20344 |
0.15 |
| chr17_74903546_74903743 | 1.16 |
Ttc27 |
tetratricopeptide repeat domain 27 |
41723 |
0.12 |
| chr16_57496238_57496389 | 1.16 |
Filip1l |
filamin A interacting protein 1-like |
52929 |
0.14 |
| chr7_16033774_16033925 | 1.16 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
14072 |
0.12 |
| chr12_69296510_69297663 | 1.16 |
Klhdc2 |
kelch domain containing 2 |
331 |
0.79 |
| chr3_145684799_145685199 | 1.15 |
Gm17501 |
predicted gene, 17501 |
32905 |
0.14 |
| chr7_143805533_143805684 | 1.14 |
Nadsyn1 |
NAD synthetase 1 |
6767 |
0.12 |
| chr9_118596882_118597033 | 1.14 |
Gm45897 |
predicted gene 45897 |
6762 |
0.16 |
| chr15_78983825_78984346 | 1.12 |
Triobp |
TRIO and F-actin binding protein |
380 |
0.7 |
| chr10_74933997_74934174 | 1.11 |
Gnaz |
guanine nucleotide binding protein, alpha z subunit |
33092 |
0.17 |
| chr10_14210602_14210753 | 1.11 |
Gm48843 |
predicted gene, 48843 |
13662 |
0.14 |
| chrX_71049982_71050218 | 1.10 |
Mamld1 |
mastermind-like domain containing 1 |
140 |
0.97 |
| chr17_28023058_28023248 | 1.08 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
15808 |
0.11 |
| chr8_103235765_103235916 | 1.08 |
Gm32531 |
predicted gene, 32531 |
35551 |
0.21 |
| chr13_92903779_92903930 | 1.08 |
Gm31630 |
predicted gene, 31630 |
5568 |
0.26 |
| chr13_31958881_31959032 | 1.07 |
Gm35732 |
predicted gene, 35732 |
12130 |
0.24 |
| chr19_49539550_49540009 | 1.07 |
Gm50444 |
predicted gene, 50444 |
14239 |
0.3 |
| chr17_66739062_66739219 | 1.07 |
Gm47471 |
predicted gene, 47471 |
386 |
0.82 |
| chr1_22579584_22579735 | 1.06 |
Rims1 |
regulating synaptic membrane exocytosis 1 |
67130 |
0.14 |
| chr14_28509611_28510106 | 1.06 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
760 |
0.59 |
| chr5_112790749_112791049 | 1.05 |
1700095B10Rik |
RIKEN cDNA 1700095B10 gene |
2393 |
0.31 |
| chr18_37835264_37836303 | 1.05 |
Gm29994 |
predicted gene, 29994 |
5583 |
0.07 |
| chr13_78168220_78168407 | 1.04 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
3317 |
0.18 |
| chr3_41366940_41367146 | 1.04 |
1700052H01Rik |
RIKEN cDNA 1700052H01 gene |
583 |
0.78 |
| chr7_29859673_29860513 | 1.02 |
Zfp420 |
zinc finger protein 420 |
52 |
0.94 |
| chr9_60837244_60837770 | 1.01 |
Gm9869 |
predicted gene 9869 |
692 |
0.7 |
| chr11_76353037_76353188 | 1.00 |
Nxn |
nucleoredoxin |
45956 |
0.12 |
| chr19_7191711_7192076 | 1.00 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
8571 |
0.13 |
| chr10_80371487_80371813 | 0.98 |
Gm15123 |
predicted gene 15123 |
248 |
0.5 |
| chr14_65023825_65023976 | 0.98 |
Ints9 |
integrator complex subunit 9 |
73587 |
0.08 |
| chr5_74105570_74105721 | 0.97 |
Snora26 |
small nucleolar RNA, H/ACA box 26 |
12115 |
0.12 |
| chr16_92300765_92301981 | 0.97 |
Smim11 |
small integral membrane protein 11 |
77 |
0.95 |
| chr9_57949183_57949334 | 0.97 |
Sema7a |
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
9145 |
0.15 |
| chr7_16891916_16892571 | 0.97 |
Gng8 |
guanine nucleotide binding protein (G protein), gamma 8 |
28 |
0.93 |
| chr12_36314056_36314241 | 0.96 |
Sostdc1 |
sclerostin domain containing 1 |
9 |
0.97 |
| chr14_50998390_50998679 | 0.95 |
Gm49038 |
predicted gene, 49038 |
1459 |
0.21 |
| chr11_78708786_78708937 | 0.95 |
Gm11197 |
predicted gene 11197 |
900 |
0.51 |
| chr12_100982252_100982605 | 0.94 |
Ccdc88c |
coiled-coil domain containing 88C |
71 |
0.96 |
| chr7_16433468_16433627 | 0.94 |
Gm45510 |
predicted gene 45510 |
16926 |
0.09 |
| chr16_13825400_13825555 | 0.92 |
Ntan1 |
N-terminal Asn amidase |
1971 |
0.23 |
| chr1_89102276_89102427 | 0.92 |
Gm38312 |
predicted gene, 38312 |
5278 |
0.22 |
| chr18_4008502_4008856 | 0.92 |
Gm7378 |
predicted gene 7378 |
1577 |
0.47 |
| chr6_87099926_87100303 | 0.92 |
D6Ertd527e |
DNA segment, Chr 6, ERATO Doi 527, expressed |
4632 |
0.18 |
| chr10_119024409_119024744 | 0.92 |
Gm47461 |
predicted gene, 47461 |
2906 |
0.23 |
| chr3_23592711_23592862 | 0.91 |
4930412E21Rik |
RIKEN cDNA 4930412E21 gene |
131736 |
0.06 |
| chr15_93710602_93710763 | 0.91 |
Gm41386 |
predicted gene, 41386 |
2087 |
0.33 |
| chr4_128451391_128451542 | 0.90 |
Csmd2 |
CUB and Sushi multiple domains 2 |
25688 |
0.23 |
| chr17_28428928_28429079 | 0.90 |
Fkbp5 |
FK506 binding protein 5 |
374 |
0.75 |
| chr1_52521544_52521856 | 0.89 |
Rps27a-ps1 |
ribosomal protein S27A, pseudogene 1 |
539 |
0.61 |
| chr9_43021695_43021846 | 0.89 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
4890 |
0.23 |
| chr13_112278397_112278548 | 0.89 |
Ankrd55 |
ankyrin repeat domain 55 |
9979 |
0.18 |
| chr10_42027737_42027888 | 0.88 |
Armc2 |
armadillo repeat containing 2 |
9370 |
0.19 |
| chr2_115857270_115857754 | 0.87 |
Meis2 |
Meis homeobox 2 |
11355 |
0.3 |
| chr5_44922248_44922762 | 0.87 |
4930431F12Rik |
RIKEN cDNA 4930431F12 gene |
43695 |
0.13 |
| chr13_56609197_56609380 | 0.86 |
Tgfbi |
transforming growth factor, beta induced |
235 |
0.93 |
| chr14_52310967_52311148 | 0.86 |
Sall2 |
spalt like transcription factor 2 |
5266 |
0.09 |
| chr10_95112565_95112841 | 0.85 |
Gm48868 |
predicted gene, 48868 |
3676 |
0.21 |
| chr2_110949973_110950403 | 0.85 |
Ano3 |
anoctamin 3 |
184 |
0.96 |
| chr5_77126733_77126884 | 0.84 |
Gm15831 |
predicted gene 15831 |
4460 |
0.15 |
| chr12_79574493_79574791 | 0.83 |
Rad51b |
RAD51 paralog B |
247289 |
0.02 |
| chr7_111694371_111694545 | 0.83 |
Galnt18 |
polypeptide N-acetylgalactosaminyltransferase 18 |
85519 |
0.09 |
| chr16_33256241_33256392 | 0.82 |
Snx4 |
sorting nexin 4 |
4872 |
0.23 |
| chr1_156669369_156669520 | 0.82 |
Tor3a |
torsin family 3, member A |
2714 |
0.22 |
| chr15_25957234_25957385 | 0.82 |
Mir7212 |
microRNA 7212 |
9082 |
0.19 |
| chr11_103814942_103815100 | 0.79 |
Nsf |
N-ethylmaleimide sensitive fusion protein |
9079 |
0.18 |
| chr7_140081805_140082771 | 0.79 |
Caly |
calcyon neuron-specific vesicular protein |
20 |
0.95 |
| chr18_75599748_75599950 | 0.79 |
Ctif |
CBP80/20-dependent translation initiation factor |
37424 |
0.19 |
| chr10_121227999_121228457 | 0.78 |
Gm35404 |
predicted gene, 35404 |
14774 |
0.15 |
| chr9_107698185_107698416 | 0.78 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
5586 |
0.1 |
| chr18_60627283_60627434 | 0.78 |
Synpo |
synaptopodin |
2495 |
0.26 |
| chr4_8963364_8963646 | 0.78 |
Rps18-ps2 |
ribosomal protein S18, pseudogene 2 |
22246 |
0.26 |
| chr2_141299301_141299452 | 0.77 |
Macrod2os1 |
mono-ADP ribosylhydrolase 2, opposite strand 1 |
12385 |
0.28 |
Network of associatons between targets according to the STRING database.
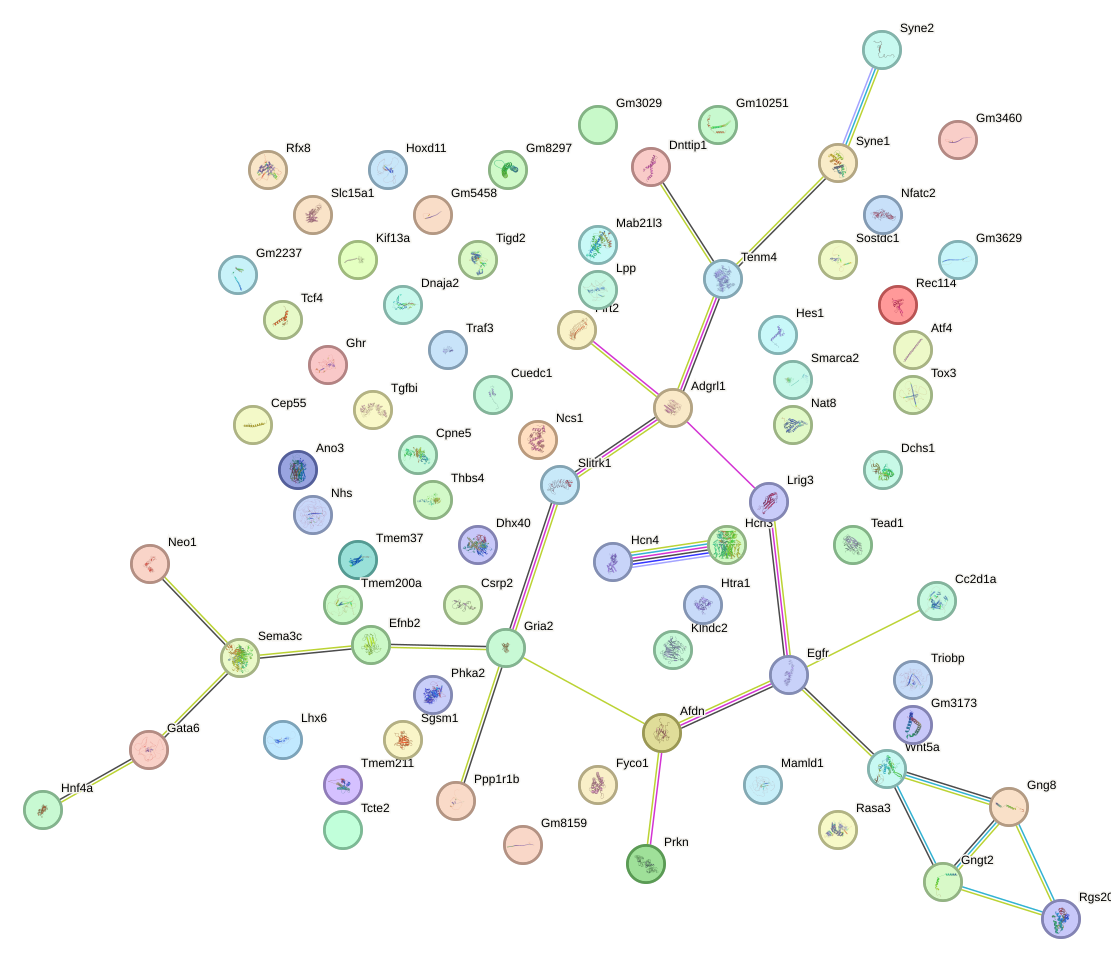
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.4 | 1.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 1.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 2.4 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.4 | 1.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 0.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 0.9 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 0.5 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.2 | 0.9 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 0.4 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.2 | 1.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.7 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.2 | 0.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 2.5 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 2.0 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.5 | GO:1904672 | regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072039) negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072040) mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:1901145) regulation of somatic stem cell population maintenance(GO:1904672) negative regulation of somatic stem cell population maintenance(GO:1904673) |
| 0.1 | 1.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.3 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.2 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.1 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.2 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.2 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 | 0.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 1.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.8 | GO:0051956 | negative regulation of amino acid transport(GO:0051956) |
| 0.1 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 1.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.5 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.2 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0060947 | cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0060413 | atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:0032341 | aldosterone metabolic process(GO:0032341) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.0 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis(GO:1902338) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.0 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.3 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0046490 | isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 5.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 0.8 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.2 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 0.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.6 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 | 0.1 | GO:0005217 | intracellular ligand-gated ion channel activity(GO:0005217) |
| 0.1 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.2 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.1 | 0.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.8 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.5 | GO:0043806 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) S-succinyltransferase activity(GO:0016751) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.0 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME CELL CELL COMMUNICATION | Genes involved in Cell-Cell communication |
| 0.1 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |