Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
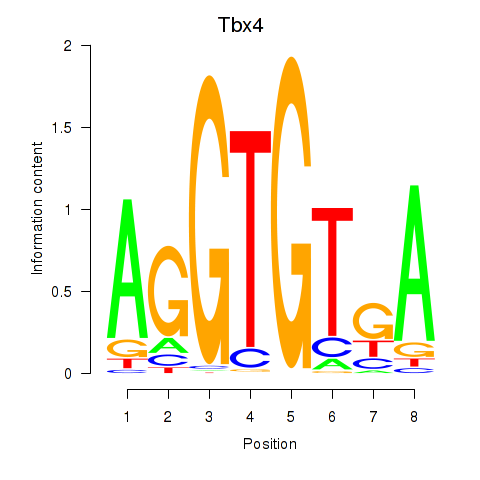
Results for Tbx4
Z-value: 0.51
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSMUSG00000000094.6 | Tbx4 |
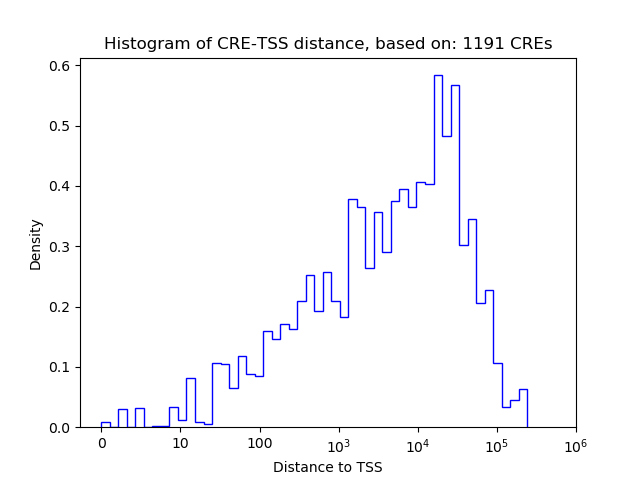
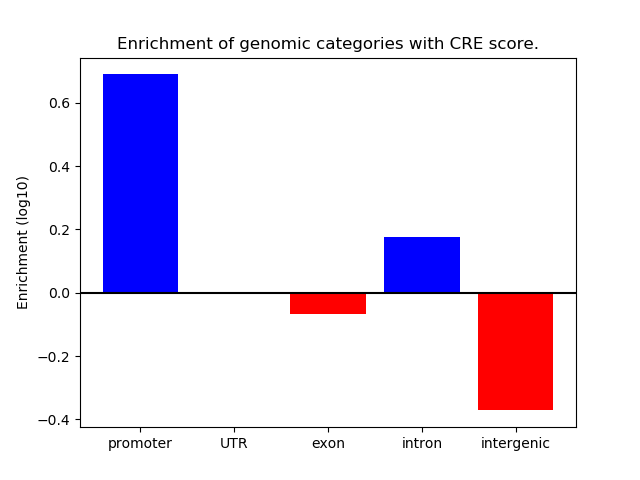
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Tbx4 | chr11_85917179_85917330 | 20489 | 0.163393 | 0.45 | 4.9e-04 | Click! |
| Tbx4 | chr11_85917416_85917567 | 20726 | 0.162977 | 0.33 | 1.4e-02 | Click! |
| Tbx4 | chr11_85918029_85918180 | 21339 | 0.161888 | 0.30 | 2.6e-02 | Click! |
| Tbx4 | chr11_85876616_85876787 | 9721 | 0.153180 | 0.29 | 3.5e-02 | Click! |
| Tbx4 | chr11_85888793_85888944 | 1195 | 0.446321 | -0.22 | 1.0e-01 | Click! |
Activity of the Tbx4 motif across conditions
Conditions sorted by the z-value of the Tbx4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
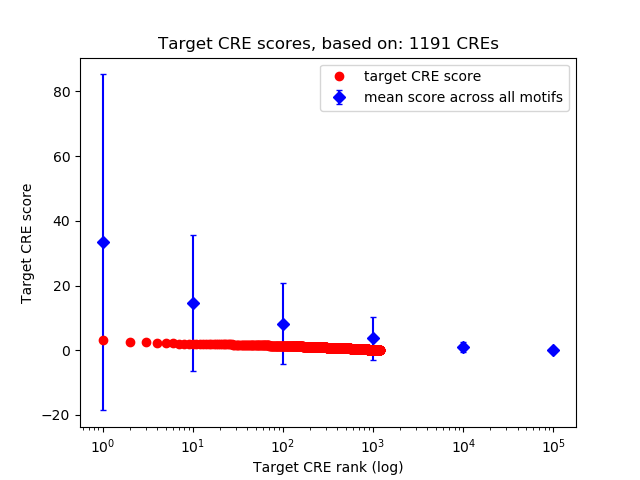
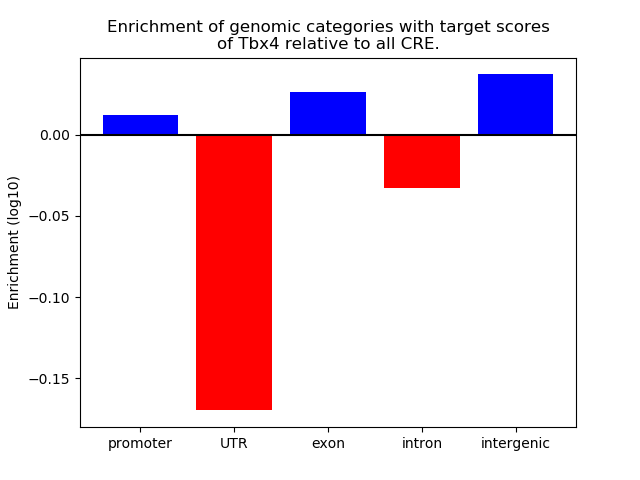
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_77207847_77208140 | 3.03 |
Spink2 |
serine peptidase inhibitor, Kazal type 2 |
3212 |
0.19 |
| chr7_24316125_24316983 | 2.68 |
Zfp94 |
zinc finger protein 94 |
28 |
0.92 |
| chr6_55580187_55580359 | 2.44 |
Gm44352 |
predicted gene, 44352 |
46767 |
0.15 |
| chr1_6730002_6730187 | 2.30 |
St18 |
suppression of tumorigenicity 18 |
7 |
0.99 |
| chr16_33605736_33606716 | 2.15 |
Slc12a8 |
solute carrier family 12 (potassium/chloride transporters), member 8 |
9599 |
0.26 |
| chr19_53449312_53449723 | 2.08 |
Mirt1 |
myocardial infarction associated transcript 1 |
1974 |
0.25 |
| chr15_66286727_66287219 | 2.04 |
Gm27242 |
predicted gene 27242 |
56 |
0.82 |
| chr5_22262595_22262780 | 2.04 |
Gm16113 |
predicted gene 16113 |
18895 |
0.17 |
| chr1_85588537_85589247 | 2.02 |
Sp110 |
Sp110 nuclear body protein |
299 |
0.81 |
| chr10_81472309_81472908 | 2.01 |
Celf5 |
CUGBP, Elav-like family member 5 |
394 |
0.64 |
| chr9_121606831_121606982 | 2.00 |
Lyzl4os |
lysozyme-like 4, opposite strand |
4062 |
0.16 |
| chr7_30291145_30292058 | 1.91 |
Clip3 |
CAP-GLY domain containing linker protein 3 |
127 |
0.89 |
| chr3_145890165_145890867 | 1.89 |
Rpl36a-ps2 |
ribosomal protein L36A, pseudogene 2 |
11496 |
0.18 |
| chr12_3238767_3239202 | 1.89 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
2373 |
0.24 |
| chr13_71690114_71690396 | 1.88 |
Gm47812 |
predicted gene, 47812 |
65227 |
0.12 |
| chr3_52784594_52784745 | 1.85 |
Gm30292 |
predicted gene, 30292 |
13116 |
0.19 |
| chr4_65838866_65839193 | 1.84 |
Trim32 |
tripartite motif-containing 32 |
233780 |
0.02 |
| chr3_89233602_89234072 | 1.84 |
Muc1 |
mucin 1, transmembrane |
2366 |
0.09 |
| chr15_75566838_75567525 | 1.83 |
Ly6h |
lymphocyte antigen 6 complex, locus H |
35 |
0.96 |
| chr9_58111541_58111738 | 1.81 |
Ccdc33 |
coiled-coil domain containing 33 |
2721 |
0.19 |
| chr13_6307436_6307642 | 1.80 |
Gm35615 |
predicted gene, 35615 |
12825 |
0.23 |
| chr9_61101873_61102054 | 1.80 |
4933433G08Rik |
RIKEN cDNA 4933433G08 gene |
3422 |
0.18 |
| chr8_52571597_52571748 | 1.79 |
Gm45673 |
predicted gene 45673 |
16958 |
0.29 |
| chr12_109010051_109010202 | 1.79 |
Wdr25 |
WD repeat domain 25 |
10676 |
0.14 |
| chr1_153641449_153641897 | 1.78 |
Rgs8 |
regulator of G-protein signaling 8 |
11352 |
0.15 |
| chr3_139885937_139886924 | 1.78 |
Gm43678 |
predicted gene 43678 |
73666 |
0.11 |
| chr4_136786990_136787145 | 1.77 |
Ephb2 |
Eph receptor B2 |
48776 |
0.12 |
| chr13_107542096_107542247 | 1.76 |
Gm32004 |
predicted gene, 32004 |
23152 |
0.2 |
| chr18_60925808_60926037 | 1.73 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
197 |
0.92 |
| chr5_15850216_15850406 | 1.72 |
Gm42453 |
predicted gene 42453 |
19936 |
0.14 |
| chr17_52602043_52602770 | 1.71 |
Gm27217 |
predicted gene 27217 |
254 |
0.55 |
| chr6_55920965_55921278 | 1.71 |
Itprid1 |
ITPR interacting domain containing 1 |
34296 |
0.21 |
| chr14_79773765_79773944 | 1.71 |
Pcdh8 |
protocadherin 8 |
2542 |
0.22 |
| chr9_94610676_94610827 | 1.71 |
Gm39404 |
predicted gene, 39404 |
18555 |
0.17 |
| chr17_65961618_65962124 | 1.71 |
Ankrd12 |
ankyrin repeat domain 12 |
10521 |
0.14 |
| chr8_71670876_71671939 | 1.70 |
Unc13a |
unc-13 homolog A |
329 |
0.75 |
| chr7_51662439_51662822 | 1.70 |
Gm45072 |
predicted gene 45072 |
30905 |
0.14 |
| chr19_14436131_14436458 | 1.67 |
Tle4 |
transducin-like enhancer of split 4 |
159245 |
0.04 |
| chr5_150905805_150906370 | 1.65 |
Gm43298 |
predicted gene 43298 |
23445 |
0.17 |
| chr10_17203245_17203406 | 1.65 |
Gm25382 |
predicted gene, 25382 |
86521 |
0.09 |
| chr4_25408022_25408173 | 1.64 |
Gm11894 |
predicted gene 11894 |
11252 |
0.22 |
| chr3_105530950_105531126 | 1.63 |
Gm43847 |
predicted gene 43847 |
28131 |
0.18 |
| chr12_44329338_44330111 | 1.63 |
Nrcam |
neuronal cell adhesion molecule |
474 |
0.8 |
| chr12_26886622_26886951 | 1.62 |
4933409F18Rik |
RIKEN cDNA 4933409F18 gene |
51913 |
0.18 |
| chr4_24966254_24966983 | 1.62 |
C230012O17Rik |
RIKEN cDNA C230012O17 gene |
2 |
0.49 |
| chr11_98284318_98284581 | 1.60 |
Gm20644 |
predicted gene 20644 |
933 |
0.39 |
| chr3_80878660_80878837 | 1.60 |
Glrb |
glycine receptor, beta subunit |
21493 |
0.21 |
| chr1_171342895_171343046 | 1.59 |
Nit1 |
nitrilase 1 |
1497 |
0.15 |
| chr13_104914342_104914493 | 1.59 |
Gm4938 |
predicted pseudogene 4938 |
4363 |
0.27 |
| chr7_129664007_129664606 | 1.59 |
Gm33248 |
predicted gene, 33248 |
3099 |
0.26 |
| chr2_152048577_152049360 | 1.58 |
AA387200 |
expressed sequence AA387200 |
27840 |
0.11 |
| chr18_72949026_72949703 | 1.56 |
Gm31908 |
predicted gene, 31908 |
28274 |
0.23 |
| chr3_17319141_17319707 | 1.56 |
Gm30340 |
predicted gene, 30340 |
15001 |
0.21 |
| chr9_44721587_44722104 | 1.54 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
421 |
0.66 |
| chr15_10421870_10422041 | 1.53 |
Dnajc21 |
DnaJ heat shock protein family (Hsp40) member C21 |
28037 |
0.15 |
| chr17_34398184_34398665 | 1.53 |
Gm15320 |
predicted gene 15320 |
262 |
0.58 |
| chr16_91962945_91963627 | 1.52 |
Gm27773 |
predicted gene, 27773 |
19138 |
0.1 |
| chr13_93072907_93073474 | 1.50 |
Gm4814 |
predicted gene 4814 |
114 |
0.97 |
| chr16_35592451_35593042 | 1.50 |
Gm5963 |
predicted pseudogene 5963 |
19190 |
0.18 |
| chr9_117873123_117873274 | 1.50 |
Gm26614 |
predicted gene, 26614 |
54 |
0.9 |
| chr8_45110316_45110467 | 1.50 |
Gm30504 |
predicted gene, 30504 |
90 |
0.97 |
| chr16_44665571_44665978 | 1.50 |
Nepro |
nucleolus and neural progenitor protein |
58527 |
0.1 |
| chr9_84032652_84032865 | 1.49 |
Bckdhb |
branched chain ketoacid dehydrogenase E1, beta polypeptide |
3082 |
0.26 |
| chr18_26847440_26847591 | 1.49 |
Gm49976 |
predicted gene, 49976 |
70582 |
0.12 |
| chr13_12503393_12503759 | 1.49 |
Edaradd |
EDAR (ectodysplasin-A receptor)-associated death domain |
16862 |
0.14 |
| chr12_11455707_11456527 | 1.49 |
Rad51ap2 |
RAD51 associated protein 2 |
38 |
0.97 |
| chr10_84415077_84415379 | 1.48 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
22840 |
0.15 |
| chr17_93201885_93202105 | 1.48 |
Adcyap1 |
adenylate cyclase activating polypeptide 1 |
81 |
0.97 |
| chr2_154421692_154421847 | 1.47 |
Snta1 |
syntrophin, acidic 1 |
13670 |
0.15 |
| chr13_15741859_15742225 | 1.47 |
Gm48408 |
predicted gene, 48408 |
28078 |
0.16 |
| chr11_20988071_20988283 | 1.45 |
Gm23681 |
predicted gene, 23681 |
50476 |
0.13 |
| chr10_80145825_80146996 | 1.45 |
Midn |
midnolin |
1862 |
0.15 |
| chr1_105843089_105843248 | 1.44 |
Tnfrsf11a |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
23298 |
0.17 |
| chr17_80483292_80483614 | 1.44 |
Sos1 |
SOS Ras/Rac guanine nucleotide exchange factor 1 |
3000 |
0.29 |
| chr13_112165997_112166183 | 1.44 |
Gm48802 |
predicted gene, 48802 |
5978 |
0.21 |
| chr13_74010757_74011079 | 1.43 |
Tppp |
tubulin polymerization promoting protein |
1499 |
0.22 |
| chr14_55115923_55116196 | 1.43 |
Jph4 |
junctophilin 4 |
507 |
0.61 |
| chr1_5018013_5019021 | 1.43 |
Rgs20 |
regulator of G-protein signaling 20 |
218 |
0.93 |
| chr6_6871537_6871723 | 1.43 |
Dlx6os1 |
distal-less homeobox 6, opposite strand 1 |
38 |
0.96 |
| chr3_34659334_34659485 | 1.43 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
3373 |
0.14 |
| chr1_56920700_56920851 | 1.42 |
Satb2 |
special AT-rich sequence binding protein 2 |
49065 |
0.13 |
| chr15_85494388_85494565 | 1.42 |
7530416G11Rik |
RIKEN cDNA 7530416G11 gene |
8751 |
0.19 |
| chr6_47239363_47240145 | 1.41 |
Cntnap2 |
contactin associated protein-like 2 |
4633 |
0.33 |
| chr10_93275246_93275449 | 1.41 |
Elk3 |
ELK3, member of ETS oncogene family |
35464 |
0.13 |
| chr6_146387163_146387325 | 1.40 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
9742 |
0.2 |
| chr13_58544429_58544580 | 1.40 |
Gm3131 |
predicted gene 3131 |
1524 |
0.39 |
| chr14_122450411_122450692 | 1.40 |
Gm5089 |
predicted gene 5089 |
564 |
0.66 |
| chr1_41605849_41606032 | 1.39 |
Gm28634 |
predicted gene 28634 |
76397 |
0.12 |
| chr2_109674717_109674868 | 1.38 |
Bdnf |
brain derived neurotrophic factor |
92 |
0.95 |
| chr14_45388403_45389548 | 1.38 |
Gnpnat1 |
glucosamine-phosphate N-acetyltransferase 1 |
14 |
0.96 |
| chr12_27084297_27084457 | 1.37 |
Gm9866 |
predicted gene 9866 |
30618 |
0.25 |
| chr4_154856178_154856666 | 1.37 |
Ttc34 |
tetratricopeptide repeat domain 34 |
222 |
0.91 |
| chr5_90588819_90589267 | 1.37 |
Gm42109 |
predicted gene, 42109 |
14 |
0.97 |
| chr12_27357279_27357647 | 1.36 |
Sox11 |
SRY (sex determining region Y)-box 11 |
14889 |
0.28 |
| chr7_4995595_4996371 | 1.35 |
Zfp579 |
zinc finger protein 579 |
46 |
0.92 |
| chr1_85279771_85280739 | 1.35 |
Gm16026 |
predicted pseudogene 16026 |
5148 |
0.14 |
| chr1_74542776_74542947 | 1.35 |
Plcd4 |
phospholipase C, delta 4 |
27 |
0.95 |
| chr9_121549119_121549291 | 1.35 |
Gm47095 |
predicted gene, 47095 |
13054 |
0.14 |
| chr3_38887381_38887941 | 1.35 |
C230034O21Rik |
RIKEN cDNA C230034O21 gene |
598 |
0.52 |
| chr6_48395472_48395677 | 1.35 |
Krba1 |
KRAB-A domain containing 1 |
12 |
0.67 |
| chr14_108709180_108709331 | 1.35 |
Rpl27a-ps2 |
ribosomal protein L27A, pseudogene 2 |
16612 |
0.28 |
| chr7_18926314_18927174 | 1.34 |
Nova2 |
NOVA alternative splicing regulator 2 |
856 |
0.42 |
| chr15_37786426_37786599 | 1.34 |
Ncald |
neurocalcin delta |
5488 |
0.19 |
| chr1_157439951_157440671 | 1.34 |
Cryzl2 |
crystallin zeta like 2 |
18266 |
0.12 |
| chr8_97476861_97477019 | 1.33 |
Gm5913 |
predicted gene 5913 |
47830 |
0.17 |
| chr15_83781833_83781984 | 1.33 |
Mpped1 |
metallophosphoesterase domain containing 1 |
1885 |
0.4 |
| chr3_30452791_30452942 | 1.33 |
Gm37024 |
predicted gene, 37024 |
35560 |
0.14 |
| chr18_68976284_68976447 | 1.32 |
4930546C10Rik |
RIKEN cDNA 4930546C10 gene |
24837 |
0.21 |
| chr7_105787328_105788039 | 1.32 |
Dchs1 |
dachsous cadherin related 1 |
29 |
0.95 |
| chr12_3891760_3892416 | 1.32 |
Dnmt3a |
DNA methyltransferase 3A |
344 |
0.86 |
| chr2_19910475_19910626 | 1.32 |
Etl4 |
enhancer trap locus 4 |
770 |
0.63 |
| chr7_25003802_25003953 | 1.31 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
2018 |
0.19 |
| chr11_82387975_82388126 | 1.31 |
Gm38577 |
predicted gene, 38577 |
710 |
0.49 |
| chr19_14240367_14240560 | 1.31 |
Gm26993 |
predicted gene, 26993 |
222092 |
0.02 |
| chr17_15248499_15248674 | 1.31 |
Gm5091 |
predicted gene 5091 |
669 |
0.71 |
| chr2_26153959_26154346 | 1.30 |
Tmem250-ps |
transmembrane protein 250, pseudogene |
13631 |
0.11 |
| chr7_63668207_63668417 | 1.30 |
Otud7a |
OTU domain containing 7A |
17497 |
0.2 |
| chr13_36736583_36736914 | 1.29 |
Nrn1 |
neuritin 1 |
1617 |
0.34 |
| chr4_22973852_22974022 | 1.29 |
1700025O08Rik |
RIKEN cDNA 1700025O08 gene |
35502 |
0.23 |
| chr1_163150570_163150773 | 1.28 |
Gm22434 |
predicted gene, 22434 |
32444 |
0.15 |
| chr18_50034876_50035293 | 1.28 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
4055 |
0.24 |
| chr11_19554563_19554970 | 1.28 |
Gm12027 |
predicted gene 12027 |
73295 |
0.11 |
| chr2_50044863_50045062 | 1.27 |
Lypd6 |
LY6/PLAUR domain containing 6 |
21467 |
0.27 |
| chr18_56774889_56775040 | 1.27 |
Gm15345 |
predicted gene 15345 |
5247 |
0.22 |
| chr7_141070009_141070863 | 1.27 |
B4galnt4 |
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
161 |
0.88 |
| chr8_12214132_12214283 | 1.27 |
A230072I06Rik |
RIKEN cDNA A230072I06 gene |
64612 |
0.1 |
| chr4_140103440_140103668 | 1.27 |
Gm13027 |
predicted gene 13027 |
28323 |
0.17 |
| chr1_137378356_137378537 | 1.26 |
Gm23534 |
predicted gene, 23534 |
11199 |
0.24 |
| chr1_85600326_85601326 | 1.26 |
Sp140 |
Sp140 nuclear body protein |
123 |
0.9 |
| chr4_117406049_117406200 | 1.26 |
Rnf220 |
ring finger protein 220 |
30667 |
0.13 |
| chr10_26827294_26827613 | 1.26 |
Arhgap18 |
Rho GTPase activating protein 18 |
4851 |
0.26 |
| chr4_46559322_46559634 | 1.26 |
Coro2a |
coronin, actin binding protein 2A |
6962 |
0.16 |
| chr4_148327377_148327851 | 1.24 |
Gm13206 |
predicted gene 13206 |
20761 |
0.14 |
| chr17_43952334_43952615 | 1.24 |
Rcan2 |
regulator of calcineurin 2 |
525 |
0.86 |
| chr8_63530326_63530477 | 1.24 |
Gm18645 |
predicted gene, 18645 |
7479 |
0.25 |
| chr2_32175074_32175462 | 1.23 |
Gm27805 |
predicted gene, 27805 |
2097 |
0.2 |
| chr18_34089514_34089665 | 1.23 |
Gm24432 |
predicted gene, 24432 |
3856 |
0.19 |
| chr3_89660407_89660568 | 1.23 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
2031 |
0.3 |
| chr11_86807230_86808078 | 1.22 |
Dhx40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
92 |
0.98 |
| chr16_67617369_67617586 | 1.22 |
Cadm2 |
cell adhesion molecule 2 |
3016 |
0.33 |
| chr10_125388906_125389759 | 1.21 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
36 |
0.95 |
| chr4_30268060_30268211 | 1.21 |
Gm11925 |
predicted gene 11925 |
74488 |
0.13 |
| chr8_123514551_123514702 | 1.21 |
Dbndd1 |
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
764 |
0.23 |
| chr6_54785459_54785618 | 1.21 |
Znrf2 |
zinc and ring finger 2 |
31378 |
0.17 |
| chr6_119341715_119341866 | 1.21 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
9228 |
0.19 |
| chr3_147064283_147064453 | 1.21 |
Gm29771 |
predicted gene, 29771 |
31816 |
0.21 |
| chr5_148265155_148265692 | 1.20 |
Mtus2 |
microtubule associated tumor suppressor candidate 2 |
78 |
0.98 |
| chr1_172203581_172203997 | 1.20 |
Pea15a |
phosphoprotein enriched in astrocytes 15A |
2908 |
0.13 |
| chr2_24761902_24762681 | 1.20 |
Cacna1b |
calcium channel, voltage-dependent, N type, alpha 1B subunit |
756 |
0.65 |
| chr17_49966143_49966321 | 1.20 |
AC133946.1 |
oxidoreductase NAD-binding domain containing 1 (OXNAD1) pseudogene |
6595 |
0.22 |
| chr1_85489813_85490173 | 1.20 |
AC147806.1 |
novel protein |
4851 |
0.14 |
| chr12_112507366_112507590 | 1.20 |
Tmem179 |
transmembrane protein 179 |
3694 |
0.2 |
| chr13_68997784_68997935 | 1.19 |
Gm48593 |
predicted gene, 48593 |
160 |
0.89 |
| chr16_41258452_41258650 | 1.18 |
Gm47276 |
predicted gene, 47276 |
40774 |
0.18 |
| chr5_112643537_112643881 | 1.18 |
4933415J04Rik |
RIKEN cDNA 4933415J04 gene |
3374 |
0.19 |
| chr12_12904775_12904974 | 1.18 |
4930519A11Rik |
RIKEN cDNA 4930519A11 gene |
324 |
0.83 |
| chr11_40601046_40601211 | 1.18 |
Gm12137 |
predicted gene 12137 |
11590 |
0.19 |
| chr9_118476459_118476610 | 1.18 |
Eomes |
eomesodermin |
1678 |
0.28 |
| chr1_132292340_132292811 | 1.18 |
Klhdc8a |
kelch domain containing 8A |
6051 |
0.12 |
| chr3_33074496_33074647 | 1.17 |
Gm19445 |
predicted gene, 19445 |
1785 |
0.3 |
| chr11_16831942_16832164 | 1.17 |
Egfros |
epidermal growth factor receptor, opposite strand |
1351 |
0.48 |
| chr1_144151888_144152189 | 1.17 |
Rgs13 |
regulator of G-protein signaling 13 |
25237 |
0.22 |
| chr9_89622321_89623673 | 1.16 |
Minar1 |
membrane integral NOTCH2 associated receptor 1 |
128 |
0.96 |
| chr19_55424034_55424281 | 1.16 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
43143 |
0.18 |
| chr4_20460440_20460613 | 1.16 |
Gm11873 |
predicted gene 11873 |
69430 |
0.13 |
| chr6_6463850_6464144 | 1.15 |
Gm20685 |
predicted gene 20685 |
19025 |
0.19 |
| chr10_81559146_81561402 | 1.15 |
Tle5 |
TLE family member 5, transcriptional modulator |
770 |
0.38 |
| chr3_143947996_143948150 | 1.14 |
Gm42707 |
predicted gene 42707 |
6325 |
0.24 |
| chr13_54884713_54885019 | 1.14 |
Gm29431 |
predicted gene 29431 |
2604 |
0.21 |
| chr19_23959939_23960407 | 1.14 |
Fam189a2 |
family with sequence similarity 189, member A2 |
16262 |
0.16 |
| chr4_98578651_98578802 | 1.14 |
Gm22625 |
predicted gene, 22625 |
20861 |
0.18 |
| chr6_107529026_107529193 | 1.13 |
Lrrn1 |
leucine rich repeat protein 1, neuronal |
659 |
0.75 |
| chr17_55971009_55971201 | 1.13 |
Shd |
src homology 2 domain-containing transforming protein D |
329 |
0.74 |
| chr6_138581835_138581986 | 1.13 |
Lmo3 |
LIM domain only 3 |
58 |
0.96 |
| chr1_85162433_85162707 | 1.12 |
Gm6264 |
predicted gene 6264 |
1707 |
0.22 |
| chr14_55520440_55521261 | 1.12 |
Nrl |
neural retina leucine zipper gene |
417 |
0.62 |
| chr10_77146904_77147516 | 1.12 |
Gm7775 |
predicted gene 7775 |
10110 |
0.18 |
| chr1_42714883_42715034 | 1.12 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
1504 |
0.32 |
| chr1_85613380_85613547 | 1.12 |
Gm10552 |
predicted gene 10552 |
1567 |
0.22 |
| chr10_30343195_30343371 | 1.11 |
Gm4780 |
predicted gene 4780 |
29635 |
0.21 |
| chr1_167119518_167119861 | 1.11 |
Gm17868 |
predicted gene, 17868 |
21466 |
0.19 |
| chr8_84814005_84814224 | 1.11 |
Dand5 |
DAN domain family member 5, BMP antagonist |
8709 |
0.08 |
| chr2_49628188_49628339 | 1.11 |
Kif5c |
kinesin family member 5C |
8965 |
0.25 |
| chr17_56020485_56020811 | 1.11 |
Gm16276 |
predicted gene 16276 |
6104 |
0.08 |
| chr4_154855497_154855648 | 1.10 |
Ttc34 |
tetratricopeptide repeat domain 34 |
628 |
0.65 |
| chr17_5286909_5287083 | 1.10 |
Gm29050 |
predicted gene 29050 |
101767 |
0.07 |
| chr1_106629505_106629752 | 1.10 |
Gm37053 |
predicted gene, 37053 |
22758 |
0.19 |
| chr3_101350461_101350612 | 1.10 |
Igsf3 |
immunoglobulin superfamily, member 3 |
26547 |
0.12 |
| chr1_176987057_176987208 | 1.10 |
Sdccag8 |
serologically defined colon cancer antigen 8 |
10877 |
0.14 |
| chr10_121830468_121830669 | 1.09 |
Gm48804 |
predicted gene, 48804 |
1490 |
0.42 |
| chr1_85095587_85095804 | 1.09 |
Gm10553 |
predicted gene 10553 |
3817 |
0.11 |
| chr2_76498827_76499126 | 1.09 |
Osbpl6 |
oxysterol binding protein-like 6 |
121 |
0.97 |
| chr3_138784741_138784929 | 1.09 |
Tspan5 |
tetraspanin 5 |
1981 |
0.36 |
| chr8_105320451_105321442 | 1.08 |
Lrrc29 |
leucine rich repeat containing 29 |
5313 |
0.07 |
| chr6_53881319_53881470 | 1.08 |
Gm22910 |
predicted gene, 22910 |
9246 |
0.21 |
| chr4_28227956_28228107 | 1.08 |
Gm11907 |
predicted gene 11907 |
125752 |
0.05 |
| chr6_135376013_135376170 | 1.08 |
Emp1 |
epithelial membrane protein 1 |
8598 |
0.17 |
| chr16_16931024_16931224 | 1.07 |
Gm46540 |
predicted gene, 46540 |
1737 |
0.22 |
| chr3_7502758_7503252 | 1.07 |
Zc2hc1a |
zinc finger, C2HC-type containing 1A |
478 |
0.68 |
| chr5_120195065_120195267 | 1.07 |
Rbm19 |
RNA binding motif protein 19 |
19441 |
0.18 |
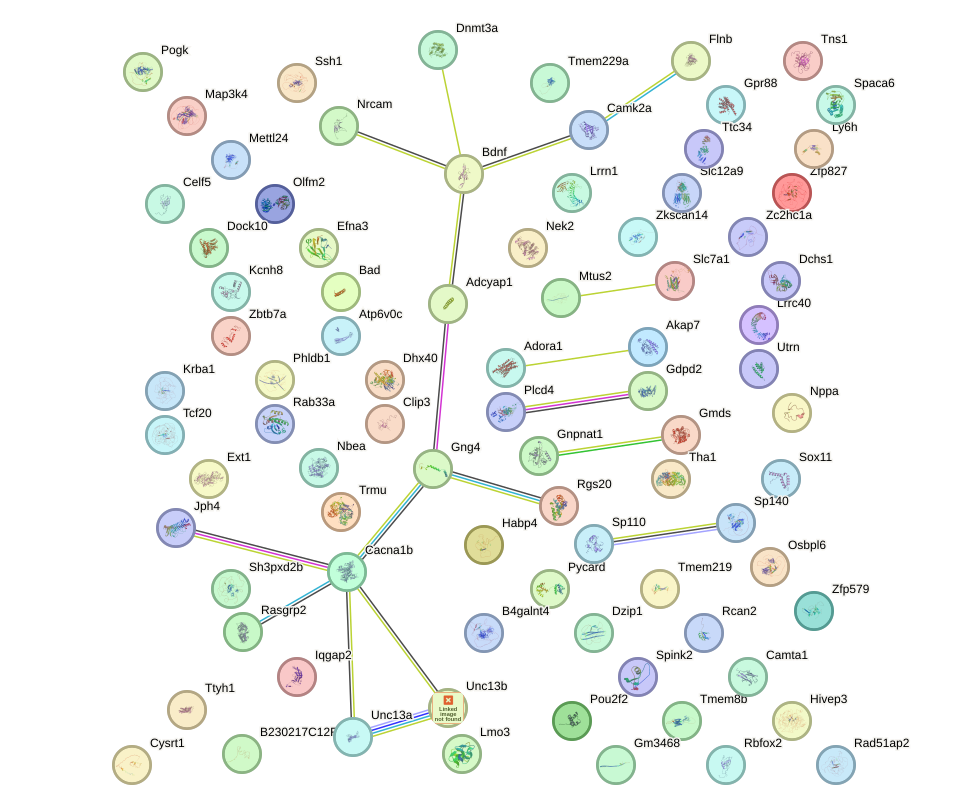
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.5 | 1.4 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.4 | 1.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 1.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 1.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 1.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 1.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.3 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.7 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.2 | 1.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.2 | 0.9 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 0.6 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.2 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 1.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 0.7 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.5 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 2.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.6 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.8 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.5 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.4 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 0.8 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.8 | GO:0098926 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.4 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.1 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.5 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.1 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.6 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.2 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.8 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.1 | 0.3 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.1 | GO:0072221 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.2 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.2 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 0.6 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.1 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.4 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.5 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0071242 | cellular response to ammonium ion(GO:0071242) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0072173 | metanephric tubule morphogenesis(GO:0072173) |
| 0.0 | 0.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:1902074 | response to salt(GO:1902074) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0071371 | cellular response to gonadotropin stimulus(GO:0071371) |
| 0.0 | 0.0 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.5 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.0 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:1903276 | regulation of sodium ion export(GO:1903273) regulation of sodium ion export from cell(GO:1903276) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0045713 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.7 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0036376 | sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0003140 | determination of left/right asymmetry in lateral mesoderm(GO:0003140) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.9 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.0 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:1903802 | amino acid import into cell(GO:1902837) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.0 | GO:0006056 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.4 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 3.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.4 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 1.6 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.4 | 1.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 0.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 0.9 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 2.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.2 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.3 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:0003905 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 1.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0043919 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.0 | GO:0004337 | geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME DNA REPLICATION | Genes involved in DNA Replication |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |