Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
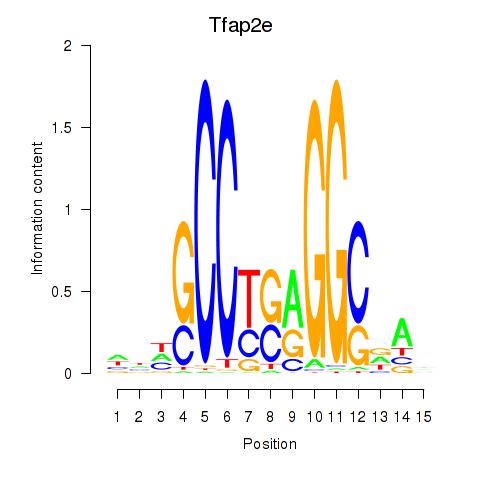
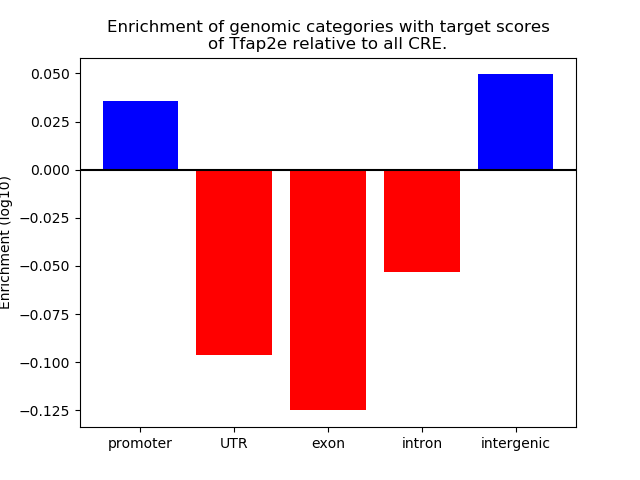
Results for Tfap2e
Z-value: 0.97
Transcription factors associated with Tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2e
|
ENSMUSG00000042477.7 | Tfap2e |
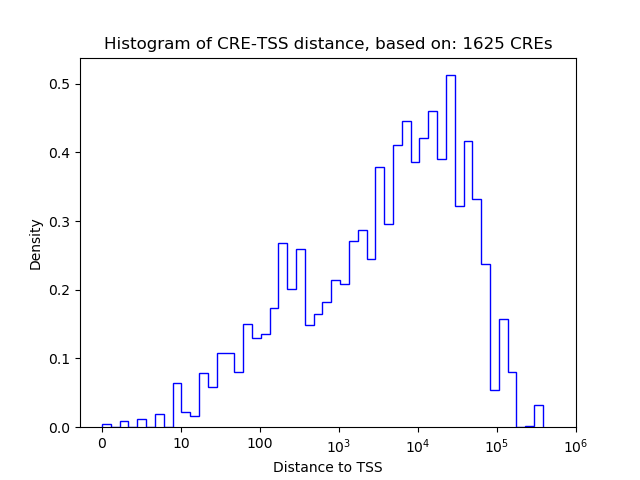
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Tfap2e | chr4_126707073_126707478 | 28965 | 0.105040 | 0.45 | 5.0e-04 | Click! |
| Tfap2e | chr4_126741576_126741746 | 5421 | 0.143386 | -0.19 | 1.6e-01 | Click! |
Activity of the Tfap2e motif across conditions
Conditions sorted by the z-value of the Tfap2e motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
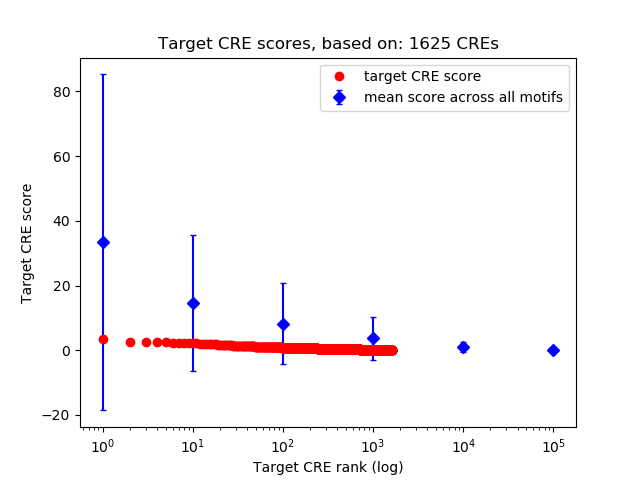
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_162221037_162221188 | 3.61 |
Dnm3os |
dynamin 3, opposite strand |
1244 |
0.37 |
| chr7_57138203_57138354 | 2.48 |
Gm18642 |
predicted gene, 18642 |
112197 |
0.07 |
| chr9_86762802_86762953 | 2.45 |
Prss35 |
protease, serine 35 |
19228 |
0.12 |
| chr10_121873065_121873455 | 2.43 |
Gm48804 |
predicted gene, 48804 |
41202 |
0.15 |
| chr4_149914094_149914250 | 2.42 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
4356 |
0.18 |
| chr18_79106086_79106237 | 2.30 |
Setbp1 |
SET binding protein 1 |
3230 |
0.37 |
| chr10_3864271_3864422 | 2.27 |
Gm16149 |
predicted gene 16149 |
5261 |
0.21 |
| chr15_10952196_10952401 | 2.22 |
C1qtnf3 |
C1q and tumor necrosis factor related protein 3 |
34 |
0.97 |
| chr12_27187507_27187842 | 2.22 |
Gm9866 |
predicted gene 9866 |
27041 |
0.24 |
| chr3_121577007_121577572 | 2.18 |
Slc44a3 |
solute carrier family 44, member 3 |
44885 |
0.11 |
| chr11_11828263_11828605 | 2.09 |
Ddc |
dopa decarboxylase |
5513 |
0.2 |
| chr14_31125276_31125695 | 2.01 |
Smim4 |
small integral membrane protein 4 |
3353 |
0.14 |
| chr13_75541571_75541722 | 1.98 |
Gm3926 |
predicted gene 3926 |
17298 |
0.15 |
| chr9_83305077_83305228 | 1.87 |
Gm46123 |
predicted gene, 46123 |
41007 |
0.13 |
| chr12_33955975_33956126 | 1.86 |
Twist1 |
twist basic helix-loop-helix transcription factor 1 |
1621 |
0.41 |
| chr5_74214849_74215000 | 1.82 |
Scfd2 |
Sec1 family domain containing 2 |
10560 |
0.15 |
| chr5_96373518_96374929 | 1.80 |
Fras1 |
Fraser extracellular matrix complex subunit 1 |
17 |
0.99 |
| chr6_22202002_22202201 | 1.78 |
Gm42573 |
predicted gene 42573 |
43833 |
0.16 |
| chr8_81260178_81260375 | 1.72 |
Gm45539 |
predicted gene 45539 |
34787 |
0.16 |
| chr13_32611765_32612081 | 1.67 |
AL645799.1 |
novel transcript |
2093 |
0.39 |
| chr11_35407713_35407933 | 1.62 |
Gm12122 |
predicted gene 12122 |
57754 |
0.14 |
| chr9_41918970_41919408 | 1.60 |
Gm40513 |
predicted gene, 40513 |
28585 |
0.14 |
| chr14_25044962_25045146 | 1.59 |
Gm47907 |
predicted gene, 47907 |
13957 |
0.17 |
| chr5_136702241_136702613 | 1.59 |
Myl10 |
myosin, light chain 10, regulatory |
4597 |
0.24 |
| chr13_18575665_18575816 | 1.54 |
Gm48829 |
predicted gene, 48829 |
69609 |
0.12 |
| chr2_21267205_21267356 | 1.53 |
Gm13378 |
predicted gene 13378 |
23175 |
0.18 |
| chr6_30169255_30169434 | 1.49 |
Mir96 |
microRNA 96 |
207 |
0.68 |
| chr4_117316934_117317085 | 1.46 |
Rnf220 |
ring finger protein 220 |
16981 |
0.13 |
| chr8_102983403_102983579 | 1.45 |
Gm8730 |
predicted pseudogene 8730 |
117693 |
0.06 |
| chr5_103029055_103029485 | 1.45 |
Mapk10 |
mitogen-activated protein kinase 10 |
2275 |
0.37 |
| chr12_118529751_118529986 | 1.43 |
D230030E09Rik |
Riken cDNA D230030E09 gene |
325 |
0.91 |
| chr4_9981765_9981916 | 1.42 |
Gm11835 |
predicted gene 11835 |
22486 |
0.19 |
| chr2_74712059_74712650 | 1.40 |
Hoxd3os1 |
homeobox D3, opposite strand 1 |
172 |
0.58 |
| chr13_111845607_111845774 | 1.39 |
Gm15325 |
predicted gene 15325 |
6932 |
0.14 |
| chr17_79145323_79145513 | 1.34 |
Gm49869 |
predicted gene, 49869 |
41674 |
0.15 |
| chr2_77816275_77817277 | 1.32 |
Zfp385b |
zinc finger protein 385B |
40 |
0.98 |
| chr16_30397261_30397963 | 1.31 |
Atp13a3 |
ATPase type 13A3 |
8363 |
0.21 |
| chr17_49816116_49816498 | 1.31 |
Gm16555 |
predicted gene 16555 |
5518 |
0.26 |
| chr10_64641714_64641867 | 1.28 |
Gm23854 |
predicted gene, 23854 |
337046 |
0.01 |
| chr17_17408850_17409001 | 1.28 |
Gm26873 |
predicted gene, 26873 |
3068 |
0.2 |
| chr13_117235503_117236043 | 1.27 |
Emb |
embigin |
7747 |
0.21 |
| chr10_52703895_52704096 | 1.26 |
Gm47624 |
predicted gene, 47624 |
10157 |
0.19 |
| chr6_147972662_147972813 | 1.24 |
Far2 |
fatty acyl CoA reductase 2 |
74522 |
0.09 |
| chr4_147343391_147343923 | 1.24 |
Zfp988 |
zinc finger protein 988 |
11250 |
0.13 |
| chr9_62721958_62722126 | 1.19 |
Itga11 |
integrin alpha 11 |
44182 |
0.14 |
| chr5_139535356_139536005 | 1.19 |
Uncx |
UNC homeobox |
7814 |
0.16 |
| chr11_87577737_87577901 | 1.16 |
Septin4 |
septin 4 |
582 |
0.66 |
| chr6_36221425_36221576 | 1.15 |
Gm43443 |
predicted gene 43443 |
32401 |
0.2 |
| chr2_174760490_174760659 | 1.15 |
Edn3 |
endothelin 3 |
45 |
0.98 |
| chr16_87341326_87341477 | 1.14 |
N6amt1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
12784 |
0.22 |
| chr13_84515013_84515539 | 1.13 |
Gm26927 |
predicted gene, 26927 |
175163 |
0.03 |
| chr3_24630114_24630286 | 1.13 |
Gm24704 |
predicted gene, 24704 |
68126 |
0.14 |
| chr18_57981702_57981894 | 1.13 |
Fbn2 |
fibrillin 2 |
45082 |
0.17 |
| chr1_83878665_83878827 | 1.10 |
4933436I20Rik |
RIKEN cDNA 4933436I20 gene |
32825 |
0.18 |
| chr18_77564045_77564233 | 1.08 |
Rnf165 |
ring finger protein 165 |
470 |
0.85 |
| chr7_108797287_108797500 | 1.08 |
Gm45232 |
predicted gene 45232 |
2390 |
0.19 |
| chr14_55716843_55717094 | 1.08 |
Rabggta |
Rab geranylgeranyl transferase, a subunit |
2178 |
0.11 |
| chr4_64047197_64047444 | 1.06 |
Tnc |
tenascin C |
305 |
0.92 |
| chr10_61866052_61866442 | 1.05 |
Col13a1 |
collagen, type XIII, alpha 1 |
166 |
0.96 |
| chr9_122285507_122285868 | 1.05 |
Ano10 |
anoctamin 10 |
8659 |
0.13 |
| chr7_19118025_19118835 | 1.05 |
Gm4969 |
predicted gene 4969 |
62 |
0.92 |
| chr5_115007642_115007807 | 1.03 |
Sppl3 |
signal peptide peptidase 3 |
3413 |
0.12 |
| chr5_49880535_49880690 | 1.03 |
Gm7988 |
predicted gene 7988 |
21690 |
0.22 |
| chr8_97233084_97233251 | 1.01 |
Gm25160 |
predicted gene, 25160 |
10633 |
0.17 |
| chr2_119196480_119196631 | 0.99 |
Gm14138 |
predicted gene 14138 |
9038 |
0.09 |
| chr4_114767178_114767329 | 0.98 |
Gm12830 |
predicted gene 12830 |
54469 |
0.11 |
| chr6_16317881_16318284 | 0.98 |
Gm3148 |
predicted gene 3148 |
80455 |
0.1 |
| chr19_56177181_56177795 | 0.98 |
Gm31912 |
predicted gene, 31912 |
71178 |
0.1 |
| chr8_121919864_121920015 | 0.98 |
Car5a |
carbonic anhydrase 5a, mitochondrial |
7535 |
0.1 |
| chr11_4308611_4309015 | 0.96 |
Gm24803 |
predicted gene, 24803 |
15442 |
0.12 |
| chr13_89741226_89741547 | 0.96 |
Vcan |
versican |
858 |
0.59 |
| chr11_7409190_7409341 | 0.95 |
Gm11986 |
predicted gene 11986 |
118891 |
0.06 |
| chr1_21461124_21461330 | 0.95 |
Gm36949 |
predicted gene, 36949 |
4210 |
0.2 |
| chr5_53266273_53266449 | 0.94 |
Smim20 |
small integral membrane protein 20 |
722 |
0.67 |
| chr7_68919196_68919350 | 0.94 |
Gm34664 |
predicted gene, 34664 |
3257 |
0.29 |
| chr17_79715582_79716163 | 0.93 |
Cyp1b1 |
cytochrome P450, family 1, subfamily b, polypeptide 1 |
811 |
0.6 |
| chr2_64325556_64325707 | 0.93 |
Gm13575 |
predicted gene 13575 |
68229 |
0.14 |
| chr3_122518107_122518786 | 0.93 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
6200 |
0.14 |
| chr19_56419505_56419787 | 0.92 |
Casp7 |
caspase 7 |
6039 |
0.19 |
| chr12_31295582_31295782 | 0.91 |
Lamb1 |
laminin B1 |
17507 |
0.13 |
| chr7_82383261_82383448 | 0.91 |
Adamtsl3 |
ADAMTS-like 3 |
25120 |
0.19 |
| chr18_60647827_60648302 | 0.89 |
Synpo |
synaptopodin |
208 |
0.94 |
| chr3_50678326_50678497 | 0.88 |
Gm37461 |
predicted gene, 37461 |
4990 |
0.24 |
| chr9_107735696_107736098 | 0.88 |
Rbm5 |
RNA binding motif protein 5 |
8337 |
0.1 |
| chrX_121232473_121232696 | 0.87 |
Gm5394 |
predicted gene 5394 |
54602 |
0.14 |
| chr1_12510492_12510833 | 0.87 |
Gm2383 |
predicted gene 2383 |
54915 |
0.14 |
| chr9_60940484_60940719 | 0.87 |
Gm47923 |
predicted gene, 47923 |
21378 |
0.18 |
| chr3_55587897_55588397 | 0.87 |
Gm43549 |
predicted gene 43549 |
5417 |
0.18 |
| chr1_82766615_82767003 | 0.87 |
Tm4sf20 |
transmembrane 4 L six family member 20 |
1652 |
0.2 |
| chr18_23877666_23877819 | 0.86 |
Gm23207 |
predicted gene, 23207 |
60371 |
0.08 |
| chr16_10192770_10193265 | 0.85 |
Atf7ip2 |
activating transcription factor 7 interacting protein 2 |
70 |
0.97 |
| chr11_78168592_78169043 | 0.85 |
Nek8 |
NIMA (never in mitosis gene a)-related expressed kinase 8 |
784 |
0.33 |
| chr1_91313342_91313536 | 0.85 |
Gm17090 |
predicted gene 17090 |
7662 |
0.11 |
| chr3_99248515_99248723 | 0.85 |
Tbx15 |
T-box 15 |
5141 |
0.17 |
| chr3_83570441_83570612 | 0.84 |
1700028M03Rik |
RIKEN cDNA 1700028M03 gene |
3596 |
0.33 |
| chr1_153096424_153096599 | 0.84 |
Nmnat2 |
nicotinamide nucleotide adenylyltransferase 2 |
3031 |
0.24 |
| chr2_27389618_27390214 | 0.84 |
Gm24049 |
predicted gene, 24049 |
11936 |
0.16 |
| chr18_74539338_74539489 | 0.83 |
1700120E14Rik |
RIKEN cDNA 1700120E14 gene |
8289 |
0.25 |
| chr18_77564959_77566098 | 0.82 |
Rnf165 |
ring finger protein 165 |
367 |
0.89 |
| chr12_100061936_100062087 | 0.82 |
9530050K03Rik |
RIKEN cDNA 9530050K03 gene |
18806 |
0.14 |
| chr9_69469113_69469304 | 0.82 |
Anxa2 |
annexin A2 |
9257 |
0.12 |
| chr12_10179432_10179754 | 0.81 |
Gm48791 |
predicted gene, 48791 |
2450 |
0.31 |
| chr6_108425133_108425318 | 0.81 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
7348 |
0.23 |
| chr17_25556611_25556991 | 0.81 |
Gm50026 |
predicted gene, 50026 |
4412 |
0.09 |
| chr1_118902149_118902552 | 0.80 |
Mir6346 |
microRNA 6346 |
51870 |
0.14 |
| chr1_171251904_171252066 | 0.80 |
Adamts4 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
131 |
0.81 |
| chr9_106464770_106466154 | 0.79 |
Gpr62 |
G protein-coupled receptor 62 |
476 |
0.6 |
| chr9_35333366_35333528 | 0.79 |
Gm33838 |
predicted gene, 33838 |
25848 |
0.11 |
| chr2_115456492_115456652 | 0.79 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
55629 |
0.14 |
| chr2_145674795_145675792 | 0.78 |
Rin2 |
Ras and Rab interactor 2 |
74 |
0.98 |
| chr1_151890040_151890576 | 0.78 |
Gm28610 |
predicted gene 28610 |
14889 |
0.19 |
| chr13_67779600_67780238 | 0.78 |
Zfp493 |
zinc finger protein 493 |
165 |
0.9 |
| chr17_29838078_29838269 | 0.77 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
1737 |
0.33 |
| chr1_15892356_15892948 | 0.77 |
Sbspon |
somatomedin B and thrombospondin, type 1 domain containing |
70 |
0.98 |
| chr11_70200228_70200379 | 0.76 |
Slc16a11 |
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
12449 |
0.07 |
| chr4_75855797_75855974 | 0.76 |
Gm11256 |
predicted gene 11256 |
1367 |
0.56 |
| chr17_14234882_14235311 | 0.76 |
Gm34567 |
predicted gene, 34567 |
14755 |
0.16 |
| chr9_103006515_103006803 | 0.75 |
Slco2a1 |
solute carrier organic anion transporter family, member 2a1 |
1343 |
0.37 |
| chr6_22288043_22288221 | 0.75 |
Wnt16 |
wingless-type MMTV integration site family, member 16 |
95 |
0.98 |
| chr1_192785556_192785974 | 0.75 |
Hhat |
hedgehog acyltransferase |
14542 |
0.15 |
| chr7_109010582_109011352 | 0.75 |
Tub |
tubby bipartite transcription factor |
142 |
0.95 |
| chr1_18416397_18416721 | 0.75 |
Defb41 |
defensin beta 41 |
151421 |
0.04 |
| chr5_110486794_110486983 | 0.75 |
Gm32996 |
predicted gene, 32996 |
4687 |
0.16 |
| chr4_151139945_151140096 | 0.75 |
Camta1 |
calmodulin binding transcription activator 1 |
436 |
0.86 |
| chr7_45682739_45683092 | 0.75 |
Ntn5 |
netrin 5 |
1107 |
0.22 |
| chr14_100619435_100619663 | 0.74 |
Gm49238 |
predicted gene, 49238 |
36212 |
0.16 |
| chr8_126538413_126539616 | 0.74 |
Gm26759 |
predicted gene, 26759 |
27956 |
0.17 |
| chr2_34982877_34983327 | 0.74 |
Hc |
hemolytic complement |
10234 |
0.12 |
| chr10_39654004_39654276 | 0.74 |
Traf3ip2 |
TRAF3 interacting protein 2 |
3585 |
0.15 |
| chr2_80447315_80447774 | 0.74 |
Frzb |
frizzled-related protein |
81 |
0.97 |
| chr5_47982936_47984535 | 0.73 |
Slit2 |
slit guidance ligand 2 |
558 |
0.73 |
| chr6_127708290_127708558 | 0.73 |
Gm42739 |
predicted gene 42739 |
55324 |
0.08 |
| chr12_112335212_112335363 | 0.73 |
Gm38123 |
predicted gene, 38123 |
14780 |
0.19 |
| chr12_13209008_13209159 | 0.73 |
Ddx1 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 |
20296 |
0.16 |
| chr1_85114233_85115011 | 0.72 |
Gm16038 |
predicted gene 16038 |
697 |
0.48 |
| chr16_18683704_18683871 | 0.72 |
Rps2-ps7 |
ribosomal protein S2, pseudogene 7 |
1571 |
0.34 |
| chr3_137977052_137977203 | 0.72 |
Dapp1 |
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
4403 |
0.15 |
| chr2_172485177_172485328 | 0.72 |
Fam209 |
family with sequence similarity 209 |
12732 |
0.13 |
| chr7_89856490_89856666 | 0.71 |
Me3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
27296 |
0.13 |
| chr7_139797631_139797782 | 0.71 |
Mir6401 |
microRNA 6401 |
18688 |
0.13 |
| chr8_125368418_125368617 | 0.71 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
124193 |
0.05 |
| chr15_77036745_77036969 | 0.71 |
Apol6 |
apolipoprotein L 6 |
7872 |
0.11 |
| chr8_87237253_87237404 | 0.71 |
Gm27169 |
predicted gene 27169 |
34302 |
0.15 |
| chr8_88451671_88451822 | 0.71 |
Gm45497 |
predicted gene 45497 |
60625 |
0.11 |
| chr17_85687855_85689066 | 0.70 |
Six2 |
sine oculis-related homeobox 2 |
186 |
0.94 |
| chr2_101592687_101592869 | 0.70 |
B230118H07Rik |
RIKEN cDNA B230118H07 gene |
7844 |
0.19 |
| chr3_67884413_67884564 | 0.70 |
Iqcj |
IQ motif containing J |
7732 |
0.23 |
| chr18_90094586_90094765 | 0.68 |
Gm6173 |
predicted gene 6173 |
75596 |
0.11 |
| chr1_52293564_52293968 | 0.68 |
Gm5975 |
predicted gene 5975 |
4671 |
0.25 |
| chr19_16841485_16841798 | 0.68 |
Gm32341 |
predicted gene, 32341 |
29451 |
0.16 |
| chr1_190979223_190979743 | 0.68 |
Vash2 |
vasohibin 2 |
187 |
0.9 |
| chr4_154781486_154781696 | 0.67 |
Ttc34 |
tetratricopeptide repeat domain 34 |
55868 |
0.1 |
| chr16_44687184_44688343 | 0.67 |
Nepro |
nucleolus and neural progenitor protein |
36538 |
0.14 |
| chr2_94729855_94730128 | 0.66 |
Gm13793 |
predicted gene 13793 |
747 |
0.75 |
| chr5_45278590_45278909 | 0.66 |
Gm43303 |
predicted gene 43303 |
26820 |
0.18 |
| chr10_57040297_57040501 | 0.66 |
Gm36827 |
predicted gene, 36827 |
58167 |
0.13 |
| chr5_104109154_104109490 | 0.66 |
Gm26703 |
predicted gene, 26703 |
31 |
0.96 |
| chr4_142674754_142674905 | 0.66 |
Gm37624 |
predicted gene, 37624 |
115979 |
0.06 |
| chr7_31076595_31076776 | 0.66 |
Fxyd3 |
FXYD domain-containing ion transport regulator 3 |
16 |
0.95 |
| chr1_118595417_118595792 | 0.65 |
Clasp1 |
CLIP associating protein 1 |
9123 |
0.17 |
| chr2_26314271_26314532 | 0.65 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
1114 |
0.34 |
| chr11_9191272_9191452 | 0.65 |
Abca13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
580 |
0.8 |
| chr18_20188319_20188643 | 0.65 |
Dsg1c |
desmoglein 1 gamma |
58859 |
0.12 |
| chr3_133877305_133877715 | 0.65 |
Gm30484 |
predicted gene, 30484 |
58992 |
0.13 |
| chr6_134128205_134128356 | 0.65 |
Gm43984 |
predicted gene, 43984 |
43679 |
0.12 |
| chr10_5922652_5922867 | 0.65 |
Rgs17 |
regulator of G-protein signaling 17 |
359 |
0.92 |
| chrX_133655989_133656226 | 0.64 |
Pcdh19 |
protocadherin 19 |
28884 |
0.24 |
| chr10_78594275_78594426 | 0.64 |
Syde1 |
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
2386 |
0.12 |
| chr2_167206316_167206467 | 0.64 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
2025 |
0.23 |
| chr14_52328141_52328337 | 0.64 |
Sall2 |
spalt like transcription factor 2 |
523 |
0.59 |
| chr2_132989928_132990079 | 0.64 |
Gm14101 |
predicted gene 14101 |
8835 |
0.16 |
| chr16_57383629_57383811 | 0.64 |
Filip1l |
filamin A interacting protein 1-like |
30443 |
0.15 |
| chrX_143664513_143664895 | 0.63 |
Pak3 |
p21 (RAC1) activated kinase 3 |
125 |
0.98 |
| chr3_97312015_97312394 | 0.63 |
Bcl9 |
B cell CLL/lymphoma 9 |
14287 |
0.18 |
| chr1_85243802_85244548 | 0.63 |
C130026I21Rik |
RIKEN cDNA C130026I21 gene |
2222 |
0.19 |
| chr12_13077810_13077961 | 0.62 |
Gm48209 |
predicted gene, 48209 |
23156 |
0.14 |
| chr18_80633674_80633825 | 0.62 |
Nfatc1 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
24774 |
0.16 |
| chr4_118037599_118037750 | 0.62 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
5792 |
0.2 |
| chr14_102981698_102983184 | 0.62 |
Mir5130 |
microRNA 5130 |
191 |
0.49 |
| chr8_23314400_23314551 | 0.61 |
Gm24335 |
predicted gene, 24335 |
24311 |
0.15 |
| chrX_169881520_169881671 | 0.61 |
Mid1 |
midline 1 |
1931 |
0.42 |
| chr2_25240744_25241709 | 0.61 |
Cysrt1 |
cysteine rich tail 1 |
1226 |
0.15 |
| chr9_8133668_8134355 | 0.61 |
Cep126 |
centrosomal protein 126 |
283 |
0.9 |
| chr2_75000578_75000751 | 0.60 |
n-R5s198 |
nuclear encoded rRNA 5S 198 |
65150 |
0.09 |
| chr19_47328251_47328788 | 0.60 |
Sh3pxd2a |
SH3 and PX domains 2A |
13768 |
0.18 |
| chr5_35609331_35609878 | 0.60 |
Acox3 |
acyl-Coenzyme A oxidase 3, pristanoyl |
495 |
0.75 |
| chr19_45796139_45796299 | 0.60 |
Kcnip2 |
Kv channel-interacting protein 2 |
515 |
0.73 |
| chr14_34936664_34936815 | 0.60 |
Mir346 |
microRNA 346 |
42130 |
0.18 |
| chr9_113479906_113480057 | 0.60 |
AU023762 |
expressed sequence AU023762 |
28374 |
0.16 |
| chr11_119514915_119515560 | 0.60 |
Endov |
endonuclease V |
13259 |
0.13 |
| chr5_129430055_129430253 | 0.60 |
Gm43001 |
predicted gene 43001 |
66034 |
0.09 |
| chr4_148918903_148919115 | 0.60 |
Gm15969 |
predicted gene 15969 |
4685 |
0.18 |
| chr4_95075494_95075799 | 0.60 |
Junos |
jun proto-oncogene, opposite strand |
23 |
0.96 |
| chr13_15464892_15465326 | 0.60 |
Gli3 |
GLI-Kruppel family member GLI3 |
1129 |
0.38 |
| chr12_107750672_107750856 | 0.59 |
4930465M20Rik |
RIKEN cDNA 4930465M20 gene |
15268 |
0.23 |
| chr9_108988525_108988676 | 0.59 |
Ucn2 |
urocortin 2 |
2590 |
0.12 |
| chr8_121468473_121468817 | 0.59 |
Gm26784 |
predicted gene, 26784 |
33665 |
0.12 |
| chr4_132469772_132470080 | 0.59 |
Med18 |
mediator complex subunit 18 |
6005 |
0.11 |
| chr12_85638862_85639281 | 0.59 |
Gm33312 |
predicted gene, 33312 |
23371 |
0.13 |
| chr2_32317120_32318698 | 0.59 |
Gm23363 |
predicted gene, 23363 |
356 |
0.45 |
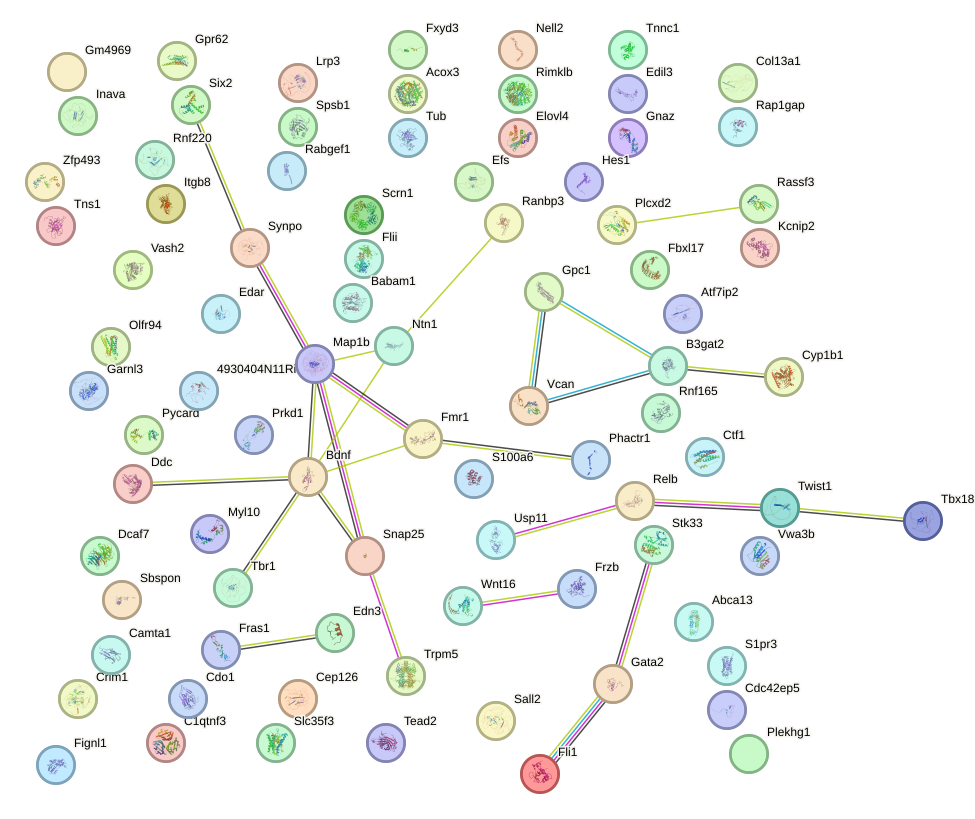
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 1.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.3 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.7 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.2 | 1.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.7 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 0.7 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.2 | 0.8 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.6 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.2 | 0.4 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.5 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.4 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 0.8 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.1 | GO:0072077 | renal vesicle morphogenesis(GO:0072077) |
| 0.1 | 0.4 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 1.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 1.8 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 1.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) |
| 0.1 | 0.3 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.3 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.8 | GO:0060384 | innervation(GO:0060384) |
| 0.1 | 0.2 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.2 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.2 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.1 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.2 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.2 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.2 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0072240 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1902805 | positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0042161 | lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.7 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.0 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0098598 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.0 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.2 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0060442 | branching involved in prostate gland morphogenesis(GO:0060442) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) |
| 0.0 | 0.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.0 | GO:0002248 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.3 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0009158 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.0 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.0 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.0 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.5 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.0 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.3 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.0 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |