Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
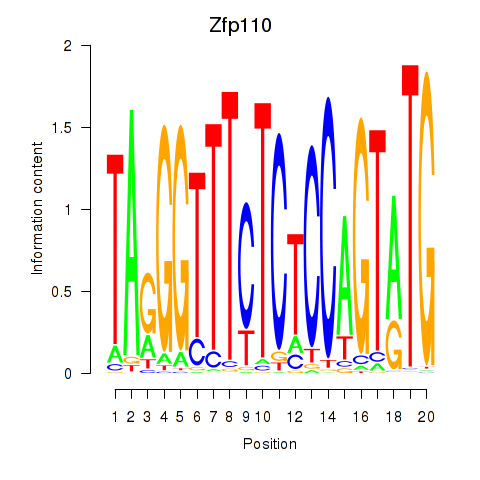
Results for Zfp110
Z-value: 0.83
Transcription factors associated with Zfp110
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp110
|
ENSMUSG00000058638.7 | Zfp110 |
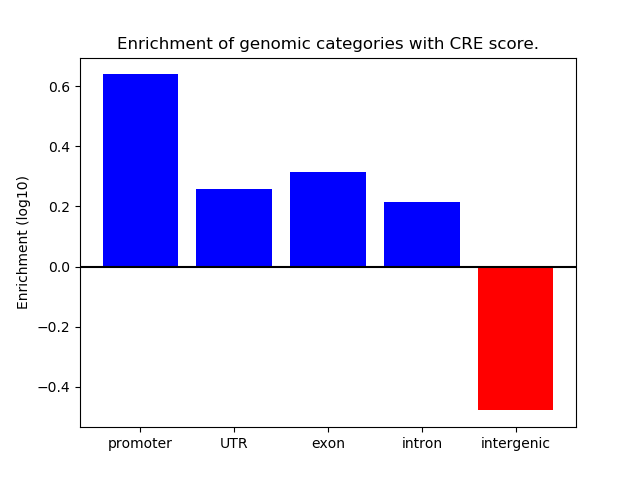
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Zfp110 | chr7_12835138_12835585 | 122 | 0.908490 | -0.17 | 2.2e-01 | Click! |
| Zfp110 | chr7_12834332_12834516 | 337 | 0.735172 | -0.07 | 6.0e-01 | Click! |
| Zfp110 | chr7_12834596_12834774 | 76 | 0.928885 | 0.06 | 6.8e-01 | Click! |
| Zfp110 | chr7_12834907_12835115 | 26 | 0.940969 | 0.02 | 8.9e-01 | Click! |
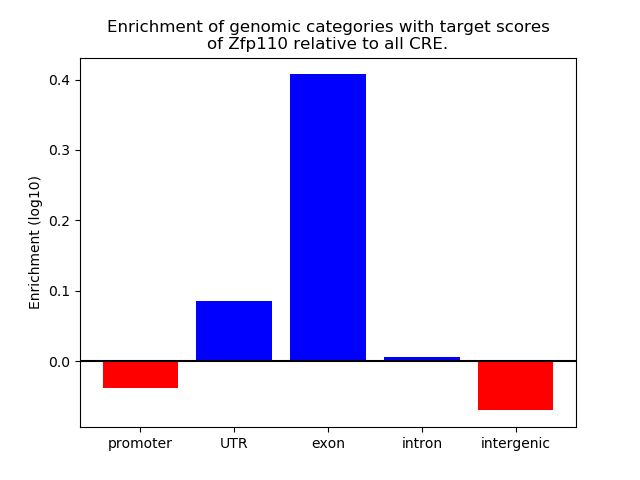
Activity of the Zfp110 motif across conditions
Conditions sorted by the z-value of the Zfp110 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
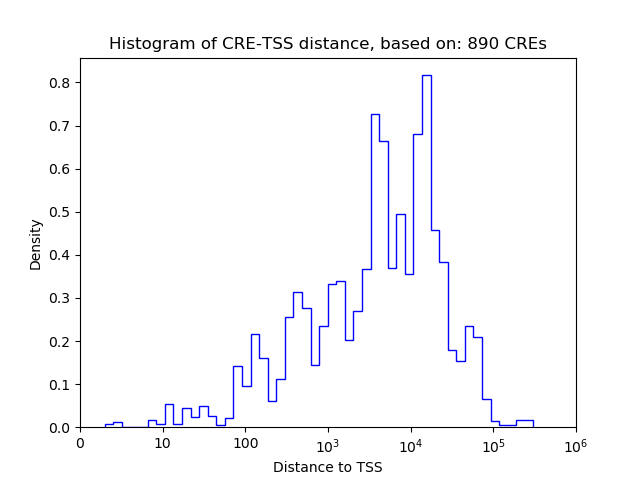
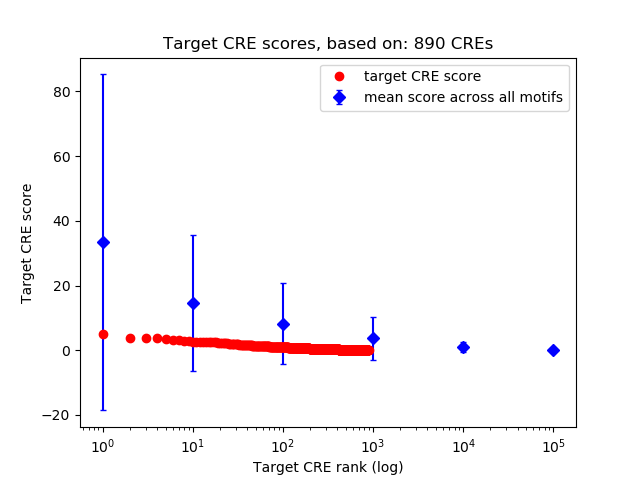
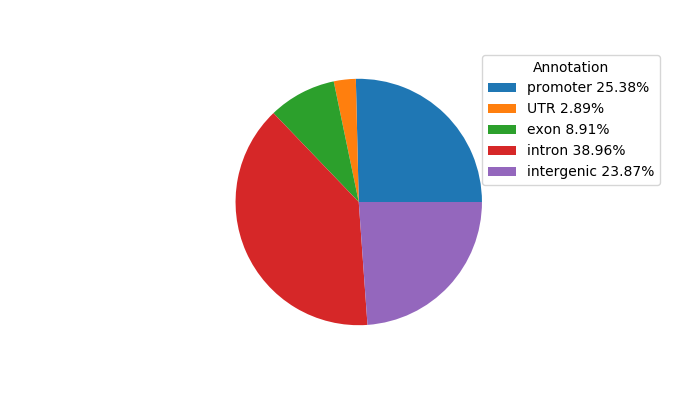
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_15544046_15544274 | 4.88 |
Gm18564 |
predicted gene, 18564 |
3656 |
0.12 |
| chr2_154632078_154632423 | 3.80 |
Gm14198 |
predicted gene 14198 |
386 |
0.76 |
| chr5_22348847_22349031 | 3.80 |
Reln |
reelin |
4237 |
0.17 |
| chr9_113915923_113916088 | 3.79 |
Ubp1 |
upstream binding protein 1 |
14929 |
0.21 |
| chr12_3618726_3618881 | 3.53 |
Dtnb |
dystrobrevin, beta |
14119 |
0.21 |
| chr17_50019206_50019365 | 3.27 |
AC133946.1 |
oxidoreductase NAD-binding domain containing 1 (OXNAD1) pseudogene |
46458 |
0.13 |
| chr17_15544423_15544600 | 3.01 |
Gm18564 |
predicted gene, 18564 |
3305 |
0.13 |
| chr17_84935739_84935899 | 2.83 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
20922 |
0.15 |
| chr5_103740202_103740381 | 2.73 |
Aff1 |
AF4/FMR2 family, member 1 |
13871 |
0.2 |
| chr2_167043032_167043339 | 2.68 |
Znfx1 |
zinc finger, NFX1-type containing 1 |
310 |
0.79 |
| chr5_75962697_75962862 | 2.62 |
Kdr |
kinase insert domain protein receptor |
15679 |
0.17 |
| chr1_161893383_161893559 | 2.56 |
Gm31925 |
predicted gene, 31925 |
1324 |
0.39 |
| chr1_23163683_23163846 | 2.48 |
Ppp1r14bl |
protein phosphatase 1, regulatory inhibitor subunit 14B like |
61511 |
0.1 |
| chr17_15544754_15544938 | 2.45 |
Gm18564 |
predicted gene, 18564 |
2970 |
0.14 |
| chr1_178779879_178780030 | 2.44 |
Kif26b |
kinesin family member 26B |
18484 |
0.21 |
| chr1_23335200_23335354 | 2.43 |
Gm20954 |
predicted gene, 20954 |
11322 |
0.15 |
| chr13_21419246_21419407 | 2.43 |
Gm50481 |
predicted gene, 50481 |
7275 |
0.07 |
| chr5_150641947_150642098 | 2.41 |
N4bp2l2 |
NEDD4 binding protein 2-like 2 |
5497 |
0.12 |
| chr3_75544411_75544575 | 2.38 |
Pdcd10 |
programmed cell death 10 |
12266 |
0.18 |
| chr17_31152594_31152769 | 2.37 |
Gm15318 |
predicted gene 15318 |
71 |
0.95 |
| chr9_98313593_98313876 | 2.33 |
Gm28530 |
predicted gene 28530 |
12084 |
0.2 |
| chr1_133825205_133825592 | 2.30 |
Gm10537 |
predicted gene 10537 |
4130 |
0.16 |
| chr19_42584309_42584471 | 2.17 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
3229 |
0.23 |
| chr2_129057908_129058064 | 2.08 |
Gm14022 |
predicted gene 14022 |
7700 |
0.11 |
| chr13_92300624_92300823 | 2.06 |
Gm20379 |
predicted gene, 20379 |
5257 |
0.22 |
| chr16_49839698_49840015 | 2.05 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
15510 |
0.24 |
| chr6_115823891_115824073 | 1.99 |
Gm44206 |
predicted gene, 44206 |
517 |
0.62 |
| chrX_73003215_73003416 | 1.99 |
Mir2137 |
microRNA 2137 |
11236 |
0.11 |
| chr2_153608221_153608636 | 1.88 |
Commd7 |
COMM domain containing 7 |
24303 |
0.15 |
| chr6_29697938_29698250 | 1.81 |
Tspan33 |
tetraspanin 33 |
3860 |
0.21 |
| chr14_26076728_26076880 | 1.79 |
Gm19014 |
predicted gene, 19014 |
3492 |
0.13 |
| chr14_25936976_25937128 | 1.78 |
Cphx1 |
cytoplasmic polyadenylated homeobox 1 |
1252 |
0.32 |
| chr14_26216351_26216502 | 1.75 |
Gm19015 |
predicted gene, 19015 |
3484 |
0.15 |
| chr7_79270956_79271107 | 1.72 |
Gm31510 |
predicted gene, 31510 |
1780 |
0.24 |
| chrX_74424134_74424310 | 1.72 |
Ikbkg |
inhibitor of kappaB kinase gamma |
312 |
0.79 |
| chr2_181419480_181419646 | 1.72 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
4825 |
0.11 |
| chr10_43291290_43291458 | 1.70 |
Gm16546 |
predicted gene 16546 |
27986 |
0.15 |
| chr9_70926847_70927150 | 1.66 |
Gm32017 |
predicted gene, 32017 |
3490 |
0.25 |
| chr19_5848083_5848533 | 1.65 |
Frmd8os |
FERM domain containing 8, opposite strand |
200 |
0.85 |
| chr13_96697612_96697763 | 1.62 |
Gm48575 |
predicted gene, 48575 |
19564 |
0.14 |
| chr7_100467548_100467843 | 1.61 |
Gm10603 |
predicted gene 10603 |
505 |
0.62 |
| chr2_173056725_173057347 | 1.57 |
Gm14453 |
predicted gene 14453 |
22456 |
0.12 |
| chr6_21790146_21790307 | 1.54 |
Tspan12 |
tetraspanin 12 |
61602 |
0.11 |
| chr17_45994256_45994531 | 1.51 |
Vegfa |
vascular endothelial growth factor A |
26979 |
0.14 |
| chr7_35601165_35601381 | 1.50 |
Ankrd27 |
ankyrin repeat domain 27 (VPS9 domain) |
153 |
0.94 |
| chr6_128845276_128845869 | 1.44 |
Gm44067 |
predicted gene, 44067 |
1387 |
0.22 |
| chr9_60269893_60270233 | 1.41 |
2010001M07Rik |
RIKEN cDNA 2010001M07 gene |
64079 |
0.12 |
| chr8_122546722_122547355 | 1.40 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
4291 |
0.11 |
| chr6_41703076_41703251 | 1.38 |
Kel |
Kell blood group |
1176 |
0.36 |
| chr12_16726590_16726741 | 1.38 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
13463 |
0.17 |
| chr1_73038176_73038388 | 1.38 |
1700027A15Rik |
RIKEN cDNA 1700027A15 gene |
13713 |
0.22 |
| chr5_122475045_122475196 | 1.36 |
Gm43359 |
predicted gene 43359 |
9879 |
0.09 |
| chr5_43052775_43052939 | 1.34 |
Gm42553 |
predicted gene 42553 |
6398 |
0.18 |
| chr5_130033922_130034092 | 1.33 |
Crcp |
calcitonin gene-related peptide-receptor component protein |
4664 |
0.14 |
| chr1_23417297_23417491 | 1.29 |
Ogfrl1 |
opioid growth factor receptor-like 1 |
19622 |
0.19 |
| chr12_80968027_80968187 | 1.29 |
Slc10a1 |
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
28 |
0.97 |
| chr7_63974223_63974578 | 1.27 |
Gm45052 |
predicted gene 45052 |
12160 |
0.13 |
| chr11_113749699_113750041 | 1.26 |
Cdc42ep4 |
CDC42 effector protein (Rho GTPase binding) 4 |
1284 |
0.38 |
| chr7_125491973_125492136 | 1.26 |
Nsmce1 |
NSE1 homolog, SMC5-SMC6 complex component |
458 |
0.81 |
| chr5_145222933_145223234 | 1.25 |
Zfp655 |
zinc finger protein 655 |
8632 |
0.1 |
| chr14_30930454_30930630 | 1.25 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
6782 |
0.11 |
| chr7_44772811_44773085 | 1.25 |
Vrk3 |
vaccinia related kinase 3 |
15962 |
0.08 |
| chr11_109615427_109615578 | 1.24 |
Gm11685 |
predicted gene 11685 |
818 |
0.56 |
| chr17_27194874_27195253 | 1.22 |
Lemd2 |
LEM domain containing 2 |
627 |
0.55 |
| chr6_91156450_91157707 | 1.21 |
Hdac11 |
histone deacetylase 11 |
86 |
0.95 |
| chr14_26595842_26595993 | 1.20 |
Dennd6a |
DENN/MADD domain containing 6A |
8754 |
0.12 |
| chr15_58592269_58592967 | 1.20 |
Fer1l6 |
fer-1-like 6 (C. elegans) |
45887 |
0.16 |
| chr2_90566138_90566391 | 1.19 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
14383 |
0.21 |
| chr18_8044181_8044768 | 1.17 |
Gm4833 |
predicted gene 4833 |
6729 |
0.28 |
| chr9_61081642_61082022 | 1.17 |
Gm34004 |
predicted gene, 34004 |
119 |
0.95 |
| chr9_88712495_88712662 | 1.13 |
Mthfsl |
5, 10-methenyltetrahydrofolate synthetase-like |
3482 |
0.13 |
| chr1_177448201_177448585 | 1.13 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
2572 |
0.22 |
| chr17_63312450_63312612 | 1.11 |
4930405O22Rik |
RIKEN cDNA 4930405O22 gene |
440 |
0.85 |
| chr7_111004187_111004476 | 1.10 |
Mrvi1 |
MRV integration site 1 |
21870 |
0.15 |
| chr2_77758483_77758775 | 1.09 |
Zfp385b |
zinc finger protein 385B |
38890 |
0.19 |
| chr7_120866301_120866479 | 1.09 |
Gm15774 |
predicted gene 15774 |
8908 |
0.13 |
| chr7_68011426_68011732 | 1.09 |
Igf1r |
insulin-like growth factor I receptor |
58752 |
0.13 |
| chr17_84183506_84183734 | 1.09 |
Gm36279 |
predicted gene, 36279 |
2136 |
0.25 |
| chr1_131125629_131125788 | 1.08 |
Dyrk3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
12537 |
0.13 |
| chr7_143930007_143930158 | 1.07 |
Mir6404 |
microRNA 6404 |
3033 |
0.25 |
| chr10_122985267_122986641 | 1.04 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
123 |
0.65 |
| chr6_98704995_98705175 | 1.04 |
Gm44196 |
predicted gene, 44196 |
7411 |
0.28 |
| chr1_133268264_133268430 | 1.03 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
1109 |
0.36 |
| chr1_13115678_13115829 | 1.02 |
Prdm14 |
PR domain containing 14 |
11410 |
0.15 |
| chr12_109436548_109437234 | 1.01 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
15932 |
0.1 |
| chrX_93631666_93632038 | 1.01 |
Pola1 |
polymerase (DNA directed), alpha 1 |
263 |
0.91 |
| chr15_9464116_9464344 | 1.01 |
Gm50456 |
predicted gene, 50456 |
11272 |
0.2 |
| chr4_45530265_45531442 | 1.01 |
Shb |
src homology 2 domain-containing transforming protein B |
523 |
0.73 |
| chr11_95858286_95858486 | 1.00 |
4833417C18Rik |
RIKEN cDNA 4833417C18 gene |
479 |
0.53 |
| chr15_80761509_80761660 | 1.00 |
Tnrc6b |
trinucleotide repeat containing 6b |
37058 |
0.13 |
| chr7_73765562_73765938 | 1.00 |
Fam174b |
family with sequence similarity 174, member B |
122 |
0.93 |
| chr2_84761983_84762134 | 0.99 |
Serping1 |
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
13070 |
0.08 |
| chr8_77044918_77045251 | 0.99 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
82929 |
0.09 |
| chr10_79897223_79897374 | 0.98 |
Med16 |
mediator complex subunit 16 |
2387 |
0.09 |
| chr7_135253282_135253611 | 0.96 |
Nps |
neuropeptide S |
5257 |
0.23 |
| chr7_81234768_81234919 | 0.94 |
Gm7180 |
predicted pseudogene 7180 |
11969 |
0.15 |
| chr2_38607587_38607812 | 0.94 |
Gm13586 |
predicted gene 13586 |
15102 |
0.12 |
| chr7_118748197_118748379 | 0.93 |
Vps35l |
VPS35 endosomal protein sorting factor like |
91 |
0.95 |
| chr13_111744433_111744584 | 0.92 |
Map3k1 |
mitogen-activated protein kinase kinase kinase 1 |
6865 |
0.15 |
| chr4_102863429_102863580 | 0.92 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
7060 |
0.27 |
| chr11_12036502_12038049 | 0.91 |
Grb10 |
growth factor receptor bound protein 10 |
126 |
0.97 |
| chr2_27470697_27470900 | 0.91 |
Brd3 |
bromodomain containing 3 |
4383 |
0.16 |
| chr5_137290779_137290972 | 0.91 |
Ache |
acetylcholinesterase |
1628 |
0.15 |
| chr6_116086165_116086341 | 0.90 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
13097 |
0.16 |
| chr10_85127631_85128502 | 0.89 |
Mterf2 |
mitochondrial transcription termination factor 2 |
39 |
0.97 |
| chr13_73443093_73443461 | 0.89 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
23920 |
0.19 |
| chr1_88248214_88248951 | 0.88 |
Mroh2a |
maestro heat-like repeat family member 2A |
9176 |
0.09 |
| chr15_86104740_86104959 | 0.88 |
Gm15722 |
predicted gene 15722 |
14789 |
0.17 |
| chr7_27794555_27794855 | 0.87 |
Zfp626 |
zinc finger protein 626 |
12491 |
0.12 |
| chr4_123663594_123663795 | 0.86 |
Macf1 |
microtubule-actin crosslinking factor 1 |
1058 |
0.4 |
| chr13_92589397_92589606 | 0.85 |
Spz1 |
spermatogenic leucine zipper 1 |
13329 |
0.19 |
| chr7_127260614_127261057 | 0.85 |
Dctpp1 |
dCTP pyrophosphatase 1 |
126 |
0.89 |
| chr5_8893870_8894350 | 0.85 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
161 |
0.94 |
| chr13_76055426_76056750 | 0.84 |
Gpr150 |
G protein-coupled receptor 150 |
908 |
0.49 |
| chr6_125560972_125561290 | 0.84 |
Vwf |
Von Willebrand factor |
5120 |
0.22 |
| chr3_105686913_105687657 | 0.81 |
Ddx20 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
159 |
0.93 |
| chr16_95929022_95929173 | 0.80 |
1600002D24Rik |
RIKEN cDNA 1600002D24 gene |
20 |
0.98 |
| chr5_36620785_36620943 | 0.80 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
391 |
0.77 |
| chr12_103367951_103368157 | 0.80 |
Gm45777 |
predicted gene 45777 |
718 |
0.5 |
| chr8_80493250_80493444 | 0.80 |
Gypa |
glycophorin A |
434 |
0.87 |
| chr4_147334693_147334844 | 0.79 |
Zfp988 |
zinc finger protein 988 |
2361 |
0.22 |
| chr16_36896797_36896959 | 0.79 |
4930565N06Rik |
RIKEN cDNA 4930565N06 gene |
311 |
0.79 |
| chr6_121336823_121336998 | 0.78 |
Slc6a12 |
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
6166 |
0.16 |
| chr19_42072579_42072730 | 0.78 |
Morn4 |
MORN repeat containing 4 |
6479 |
0.11 |
| chr5_142282102_142282295 | 0.78 |
Gm26970 |
predicted gene, 26970 |
908 |
0.71 |
| chr6_113078572_113078736 | 0.77 |
Setd5 |
SET domain containing 5 |
1015 |
0.33 |
| chrX_7966286_7966460 | 0.77 |
Gata1 |
GATA binding protein 1 |
1537 |
0.2 |
| chr7_127253701_127254925 | 0.77 |
Dctpp1 |
dCTP pyrophosphatase 1 |
6396 |
0.07 |
| chr10_78324578_78324790 | 0.77 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
3994 |
0.1 |
| chr10_118064868_118065238 | 0.76 |
5330439M10Rik |
RIKEN cDNA 5330439M10 gene |
47464 |
0.11 |
| chr7_113084925_113085076 | 0.76 |
Gm23662 |
predicted gene, 23662 |
34698 |
0.16 |
| chr2_173976360_173976532 | 0.73 |
Atp5k-ps2 |
ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E, pseudogene 2 |
8245 |
0.21 |
| chr4_130335999_130336150 | 0.73 |
Zcchc17 |
zinc finger, CCHC domain containing 17 |
2510 |
0.23 |
| chr13_15714658_15715471 | 0.72 |
Gm48408 |
predicted gene, 48408 |
55056 |
0.11 |
| chr7_110715161_110715448 | 0.72 |
Gm18907 |
predicted gene, 18907 |
1907 |
0.28 |
| chr17_27105493_27105644 | 0.72 |
Itpr3 |
inositol 1,4,5-triphosphate receptor 3 |
609 |
0.55 |
| chr18_36405636_36405997 | 0.72 |
Cystm1 |
cysteine-rich transmembrane module containing 1 |
40035 |
0.12 |
| chr17_29393661_29393812 | 0.71 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
17235 |
0.11 |
| chr2_155604070_155604221 | 0.71 |
Myh7b |
myosin, heavy chain 7B, cardiac muscle, beta |
7067 |
0.08 |
| chr14_30886332_30886731 | 0.70 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
10 |
0.96 |
| chr8_60974955_60975112 | 0.70 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
8206 |
0.15 |
| chr6_118408467_118408618 | 0.69 |
Bms1 |
BMS1, ribosome biogenesis factor |
10862 |
0.16 |
| chr5_43224358_43224509 | 0.68 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
8737 |
0.16 |
| chr7_81606016_81606167 | 0.68 |
Gm44570 |
predicted gene 44570 |
4018 |
0.14 |
| chr1_85594072_85594251 | 0.68 |
Sp110 |
Sp110 nuclear body protein |
4588 |
0.11 |
| chr17_16951328_16951952 | 0.67 |
BC002059 |
cDNA sequence BC002059 |
88 |
0.98 |
| chr1_191325205_191325584 | 0.67 |
Pacc1 |
proton activated chloride channel 1 |
518 |
0.72 |
| chr6_88251435_88251608 | 0.67 |
1700031F10Rik |
RIKEN cDNA 1700031F10 gene |
25843 |
0.12 |
| chr10_21028286_21028444 | 0.67 |
Gm10826 |
predicted gene 10826 |
5428 |
0.18 |
| chr17_48272628_48272921 | 0.66 |
Treml4 |
triggering receptor expressed on myeloid cells-like 4 |
335 |
0.82 |
| chr13_95324586_95325933 | 0.65 |
Zbed3 |
zinc finger, BED type containing 3 |
22 |
0.97 |
| chr10_127591160_127591879 | 0.65 |
Gm16217 |
predicted gene 16217 |
5029 |
0.12 |
| chr14_45653782_45654098 | 0.65 |
Ddhd1 |
DDHD domain containing 1 |
3420 |
0.14 |
| chr5_142441713_142441867 | 0.65 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
22154 |
0.17 |
| chr5_123035827_123035983 | 0.65 |
Gm43412 |
predicted gene 43412 |
95 |
0.92 |
| chr13_70866959_70867347 | 0.64 |
8030423J24Rik |
RIKEN cDNA 8030423J24 gene |
15795 |
0.19 |
| chr11_45087661_45087812 | 0.64 |
Gm12159 |
predicted gene 12159 |
29192 |
0.21 |
| chr13_104799919_104800085 | 0.64 |
Cwc27 |
CWC27 spliceosome-associated protein |
7352 |
0.21 |
| chr5_146712196_146712347 | 0.64 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
5649 |
0.19 |
| chr5_150390088_150390273 | 0.63 |
Fry |
FRY microtubule binding protein |
9473 |
0.18 |
| chr17_75551163_75552443 | 0.63 |
Fam98a |
family with sequence similarity 98, member A |
7 |
0.99 |
| chr11_3288568_3288934 | 0.62 |
Patz1 |
POZ (BTB) and AT hook containing zinc finger 1 |
123 |
0.94 |
| chr18_78344507_78344679 | 0.62 |
Gm6133 |
predicted gene 6133 |
5161 |
0.31 |
| chr1_85960049_85960213 | 0.62 |
Gpr55 |
G protein-coupled receptor 55 |
876 |
0.47 |
| chr9_120770594_120770843 | 0.61 |
Gm18101 |
predicted gene, 18101 |
15192 |
0.11 |
| chr6_67145077_67145237 | 0.61 |
A430010J10Rik |
RIKEN cDNA A430010J10 gene |
19767 |
0.15 |
| chr13_57671724_57671875 | 0.61 |
Spock1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
235788 |
0.02 |
| chr5_139368774_139368951 | 0.61 |
Mir339 |
microRNA 339 |
883 |
0.44 |
| chr6_136470792_136471312 | 0.60 |
Gm6728 |
predicted gene 6728 |
16134 |
0.12 |
| chr5_52994485_52994679 | 0.60 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
146 |
0.95 |
| chr6_87759971_87760122 | 0.59 |
Gm5879 |
predicted gene 5879 |
6158 |
0.09 |
| chr7_116288066_116288220 | 0.59 |
Gm44867 |
predicted gene 44867 |
5258 |
0.16 |
| chr5_113978054_113978205 | 0.59 |
Ssh1 |
slingshot protein phosphatase 1 |
11373 |
0.13 |
| chr11_98740095_98741203 | 0.59 |
Thra |
thyroid hormone receptor alpha |
11 |
0.95 |
| chr16_58669361_58669681 | 0.59 |
Cpox |
coproporphyrinogen oxidase |
771 |
0.54 |
| chr10_128640180_128640331 | 0.58 |
Ikzf4 |
IKAROS family zinc finger 4 |
4393 |
0.08 |
| chr11_3291684_3291986 | 0.58 |
Patz1 |
POZ (BTB) and AT hook containing zinc finger 1 |
360 |
0.79 |
| chr9_108067929_108068247 | 0.57 |
Rnf123 |
ring finger protein 123 |
5033 |
0.07 |
| chr10_75762614_75762766 | 0.57 |
Cabin1 |
calcineurin binding protein 1 |
1580 |
0.22 |
| chr12_100998964_100999115 | 0.57 |
Gm36756 |
predicted gene, 36756 |
6067 |
0.13 |
| chr2_153611372_153611523 | 0.57 |
Commd7 |
COMM domain containing 7 |
21284 |
0.15 |
| chr6_114920630_114921081 | 0.56 |
Vgll4 |
vestigial like family member 4 |
961 |
0.63 |
| chr1_88280037_88280231 | 0.56 |
Trpm8 |
transient receptor potential cation channel, subfamily M, member 8 |
2473 |
0.16 |
| chr3_103141435_103141586 | 0.55 |
Dennd2c |
DENN/MADD domain containing 2C |
542 |
0.69 |
| chr16_23517803_23517957 | 0.55 |
Gm45338 |
predicted gene 45338 |
2330 |
0.19 |
| chr7_140922127_140922278 | 0.55 |
Nlrp6 |
NLR family, pyrin domain containing 6 |
96 |
0.9 |
| chr15_74932626_74932777 | 0.55 |
Gm39556 |
predicted gene, 39556 |
3264 |
0.12 |
| chr9_71590886_71591037 | 0.55 |
Myzap |
myocardial zonula adherens protein |
1304 |
0.45 |
| chr1_181210229_181210381 | 0.54 |
Wdr26 |
WD repeat domain 26 |
1121 |
0.41 |
| chrX_11696672_11696823 | 0.54 |
Gm14513 |
predicted gene 14513 |
12623 |
0.26 |
| chr4_134883414_134883565 | 0.54 |
Rhd |
Rh blood group, D antigen |
3679 |
0.2 |
| chr7_75610098_75610263 | 0.54 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
141 |
0.96 |
| chr11_3143589_3143897 | 0.54 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
1365 |
0.27 |
| chr8_105515875_105516531 | 0.54 |
Hsd11b2 |
hydroxysteroid 11-beta dehydrogenase 2 |
2552 |
0.14 |
| chr1_137303158_137303309 | 0.53 |
1700006P03Rik |
RIKEN cDNA 1700006P03 gene |
22609 |
0.2 |
| chr16_91545542_91546220 | 0.53 |
Ifngr2 |
interferon gamma receptor 2 |
1191 |
0.3 |
| chr4_63357109_63357260 | 0.53 |
Orm3 |
orosomucoid 3 |
1022 |
0.37 |
| chr15_67174623_67174888 | 0.53 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
1957 |
0.43 |
| chr11_110997176_110997525 | 0.53 |
Kcnj16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
137 |
0.98 |
| chr2_28601544_28601695 | 0.52 |
Gm22675 |
predicted gene, 22675 |
6492 |
0.11 |
Network of associatons between targets according to the STRING database.
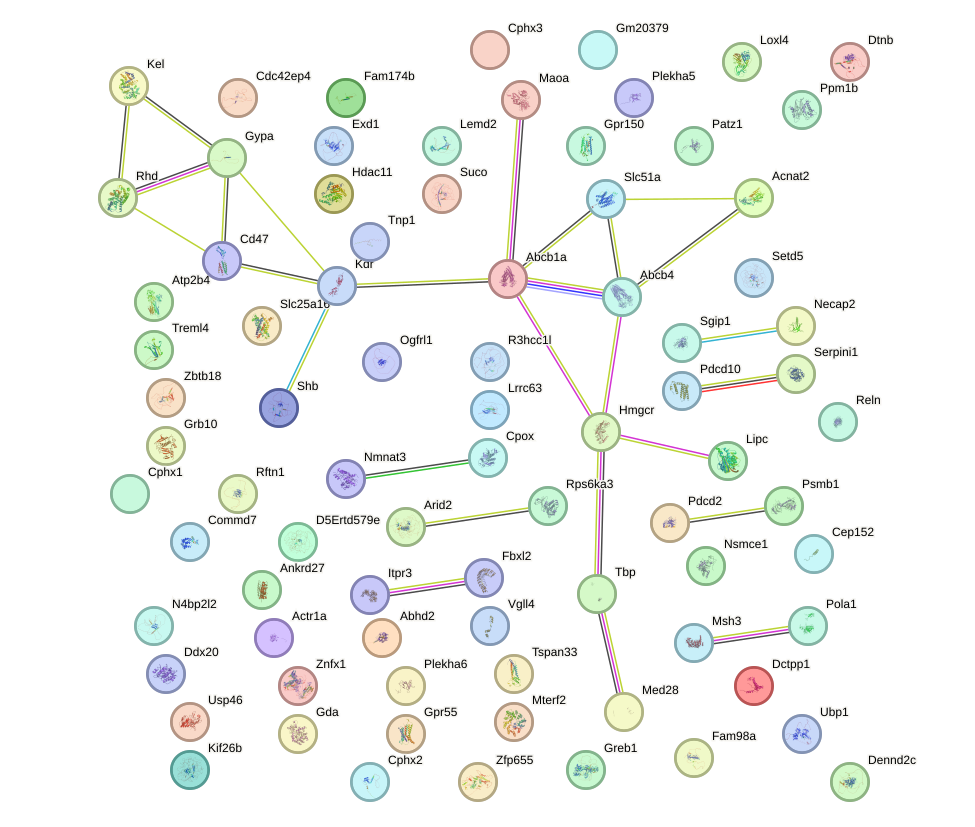
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.3 | 0.9 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.3 | 1.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.3 | 0.8 | GO:0045472 | response to ether(GO:0045472) |
| 0.2 | 0.6 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.2 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 1.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 0.7 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 0.7 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.2 | 0.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 | 0.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.7 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 0.2 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.5 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.9 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 1.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.3 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.1 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.1 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.2 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 0.2 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.8 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.1 | 0.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.1 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.2 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.2 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.3 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.5 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.0 | 1.0 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0052173 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.0 | 0.5 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.0 | 0.6 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.2 | GO:0060896 | neural plate pattern specification(GO:0060896) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.3 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0000237 | leptotene(GO:0000237) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0009813 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0022616 | DNA strand elongation(GO:0022616) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:2000416 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.0 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.0 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.0 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.0 | 0.8 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 1.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 1.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0030894 | replisome(GO:0030894) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 1.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.7 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.2 | 0.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.4 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 1.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |