Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
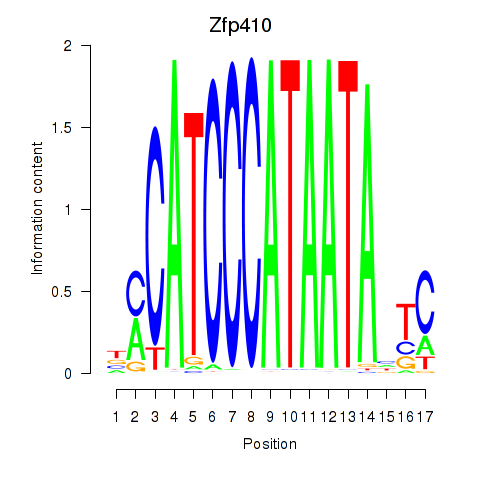
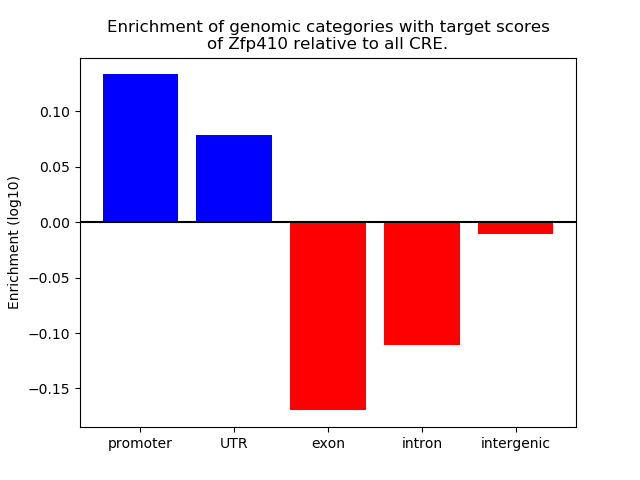
Results for Zfp410
Z-value: 1.03
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.10 | Zfp410 |
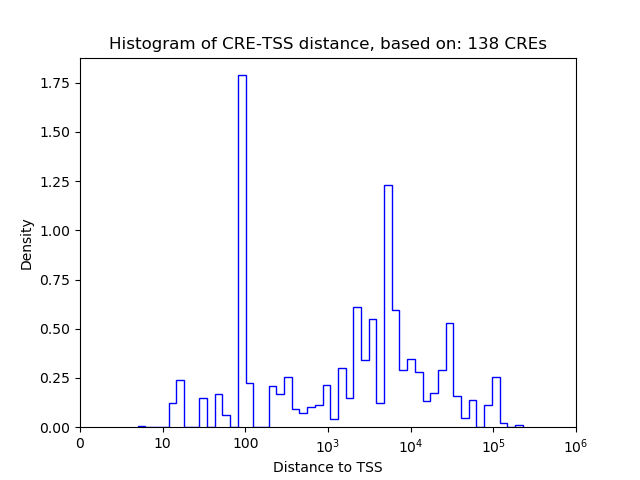
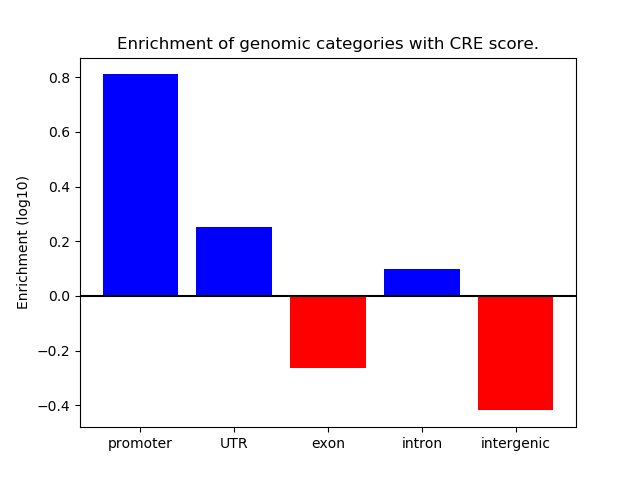
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Zfp410 | chr12_84317435_84317637 | 391 | 0.494588 | 0.52 | 5.4e-05 | Click! |
| Zfp410 | chr12_84316711_84316909 | 42 | 0.768168 | 0.43 | 9.6e-04 | Click! |
| Zfp410 | chr12_84317218_84317412 | 170 | 0.578424 | 0.42 | 1.5e-03 | Click! |
| Zfp410 | chr12_84304058_84304308 | 12669 | 0.108046 | 0.37 | 6.0e-03 | Click! |
| Zfp410 | chr12_84327310_84327462 | 54 | 0.956268 | 0.18 | 1.8e-01 | Click! |
Activity of the Zfp410 motif across conditions
Conditions sorted by the z-value of the Zfp410 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
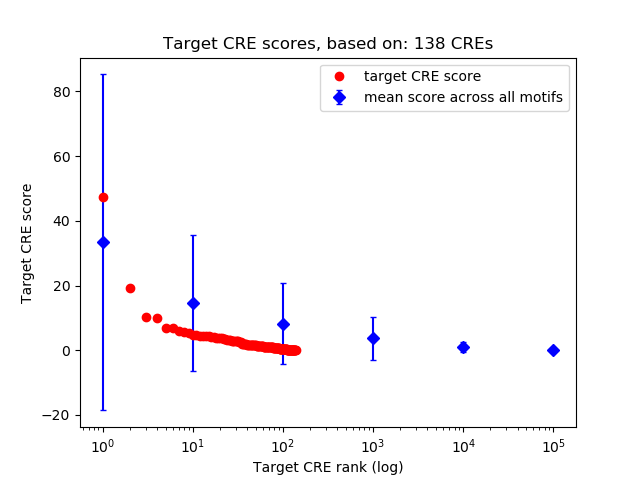
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_125095392_125097556 | 47.37 |
Chd4 |
chromodomain helicase DNA binding protein 4 |
95 |
0.84 |
| chr6_125088567_125089479 | 19.27 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
5586 |
0.08 |
| chr6_125089773_125090131 | 10.40 |
Chd4 |
chromodomain helicase DNA binding protein 4 |
6029 |
0.07 |
| chr10_60078002_60078293 | 10.07 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
28072 |
0.16 |
| chr2_153495495_153495649 | 7.01 |
4930404H24Rik |
RIKEN cDNA 4930404H24 gene |
2782 |
0.24 |
| chr15_44462509_44462660 | 6.95 |
Pkhd1l1 |
polycystic kidney and hepatic disease 1-like 1 |
5031 |
0.19 |
| chr17_36030163_36030330 | 5.92 |
H2-T23 |
histocompatibility 2, T region locus 23 |
2295 |
0.1 |
| chr6_144901597_144901788 | 5.65 |
Gm22792 |
predicted gene, 22792 |
98060 |
0.06 |
| chr7_110088063_110088458 | 5.23 |
Zfp143 |
zinc finger protein 143 |
3369 |
0.16 |
| chr4_123285054_123285219 | 4.78 |
Pabpc4 |
poly(A) binding protein, cytoplasmic 4 |
2041 |
0.17 |
| chr16_22893442_22893593 | 4.63 |
Gm30505 |
predicted gene, 30505 |
869 |
0.37 |
| chr5_103977347_103977558 | 4.51 |
Hsd17b13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
42 |
0.97 |
| chr1_131526874_131527597 | 4.46 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
112 |
0.89 |
| chr11_102368389_102368661 | 4.42 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
2322 |
0.16 |
| chr10_59347750_59347911 | 4.35 |
P4ha1 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
24462 |
0.16 |
| chr16_21788047_21788243 | 4.01 |
Ehhadh |
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
338 |
0.82 |
| chr2_30658442_30659224 | 3.97 |
Gm14486 |
predicted gene 14486 |
11075 |
0.15 |
| chr5_123142139_123142306 | 3.90 |
Setd1b |
SET domain containing 1B |
29 |
0.88 |
| chr12_111437288_111437481 | 3.90 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
5085 |
0.13 |
| chr1_31094793_31095271 | 3.78 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
1422 |
0.36 |
| chr10_56106920_56107369 | 3.64 |
Msl3l2 |
MSL3 like 2 |
227 |
0.94 |
| chr12_85704751_85704902 | 3.62 |
Gm47285 |
predicted gene, 47285 |
13737 |
0.12 |
| chr6_98589121_98589272 | 3.37 |
4930595L18Rik |
RIKEN cDNA 4930595L18 gene |
37089 |
0.21 |
| chr15_79833949_79834578 | 3.30 |
Cbx6 |
chromobox 6 |
11 |
0.5 |
| chr5_100500528_100500689 | 3.24 |
Lin54 |
lin-54 homolog (C. elegans) |
16 |
0.85 |
| chr11_119040519_119041332 | 3.16 |
Cbx8 |
chromobox 8 |
14 |
0.97 |
| chr12_12488288_12488471 | 2.98 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
95764 |
0.08 |
| chr8_110933315_110933782 | 2.95 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
1627 |
0.24 |
| chr7_103875101_103875284 | 2.93 |
Olfr66 |
olfactory receptor 66 |
7049 |
0.06 |
| chr7_64040446_64040628 | 2.89 |
Gm45054 |
predicted gene 45054 |
289 |
0.89 |
| chr6_90608041_90608201 | 2.76 |
Slc41a3 |
solute carrier family 41, member 3 |
3396 |
0.18 |
| chr14_67941339_67941490 | 2.76 |
Dock5 |
dedicator of cytokinesis 5 |
7972 |
0.25 |
| chr7_99590701_99590937 | 2.48 |
Arrb1 |
arrestin, beta 1 |
3774 |
0.14 |
| chr18_4804091_4804297 | 2.43 |
Gm10556 |
predicted gene 10556 |
8292 |
0.26 |
| chr6_32479585_32479786 | 2.06 |
Plxna4os2 |
plexin A4, opposite strand 2 |
28515 |
0.19 |
| chr5_139803949_139804524 | 2.05 |
Tmem184a |
transmembrane protein 184a |
3744 |
0.14 |
| chr17_71251382_71251788 | 2.04 |
Emilin2 |
elastin microfibril interfacer 2 |
3821 |
0.19 |
| chr12_85785503_85785771 | 1.97 |
Flvcr2 |
feline leukemia virus subgroup C cellular receptor 2 |
10807 |
0.15 |
| chr3_10316055_10316213 | 1.96 |
Impa1 |
inositol (myo)-1(or 4)-monophosphatase 1 |
518 |
0.62 |
| chr2_130577744_130577954 | 1.75 |
Oxt |
oxytocin |
1676 |
0.2 |
| chr11_86382617_86383101 | 1.73 |
Med13 |
mediator complex subunit 13 |
25257 |
0.16 |
| chr15_102103920_102104453 | 1.64 |
Tns2 |
tensin 2 |
239 |
0.87 |
| chr10_24911202_24911353 | 1.63 |
Mir6905 |
microRNA 6905 |
613 |
0.63 |
| chr17_27796564_27796715 | 1.58 |
Rps2-ps9 |
ribosomal protein S2, pseudogene 9 |
12215 |
0.12 |
| chr14_64949310_64950069 | 1.58 |
Hmbox1 |
homeobox containing 1 |
57 |
0.83 |
| chr4_135873258_135873453 | 1.56 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
215 |
0.88 |
| chr19_23687766_23687949 | 1.54 |
Ptar1 |
protein prenyltransferase alpha subunit repeat containing 1 |
113 |
0.95 |
| chr13_73448622_73448799 | 1.54 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
18487 |
0.2 |
| chr15_86110952_86111103 | 1.53 |
Gm15722 |
predicted gene 15722 |
20967 |
0.16 |
| chr7_12978404_12978717 | 1.52 |
Zfp446 |
zinc finger protein 446 |
322 |
0.74 |
| chr13_81853046_81853197 | 1.42 |
Cetn3 |
centrin 3 |
61144 |
0.11 |
| chr2_5549339_5549504 | 1.36 |
Gm13216 |
predicted gene 13216 |
54150 |
0.15 |
| chr1_156222336_156222944 | 1.35 |
Gm38113 |
predicted gene, 38113 |
4653 |
0.18 |
| chrX_150549242_150549398 | 1.34 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
1861 |
0.24 |
| chr9_88483752_88484150 | 1.30 |
Syncrip |
synaptotagmin binding, cytoplasmic RNA interacting protein |
1377 |
0.28 |
| chr14_52196866_52197101 | 1.25 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
433 |
0.47 |
| chr13_3744124_3744306 | 1.25 |
Gm47695 |
predicted gene, 47695 |
15610 |
0.15 |
| chr2_33788959_33789191 | 1.21 |
Mvb12b |
multivesicular body subunit 12B |
316 |
0.9 |
| chr7_67229002_67229160 | 1.17 |
Lysmd4 |
LysM, putative peptidoglycan-binding, domain containing 4 |
5472 |
0.17 |
| chr2_38504413_38504578 | 1.17 |
Gm44062 |
predicted gene, 44062 |
4303 |
0.14 |
| chr11_3134327_3135400 | 1.12 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
1144 |
0.38 |
| chr12_81002763_81003134 | 1.12 |
Smoc1 |
SPARC related modular calcium binding 1 |
23860 |
0.14 |
| chr4_97025797_97025948 | 1.05 |
Gm27521 |
predicted gene, 27521 |
108852 |
0.07 |
| chr10_40152341_40152498 | 1.03 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
10161 |
0.13 |
| chr8_108613044_108613195 | 1.03 |
Zfhx3 |
zinc finger homeobox 3 |
781 |
0.71 |
| chr9_57970489_57970640 | 1.01 |
Gm17322 |
predicted gene, 17322 |
27477 |
0.11 |
| chr9_73044180_73044397 | 1.01 |
Rab27a |
RAB27A, member RAS oncogene family |
566 |
0.54 |
| chr4_129671576_129671731 | 1.00 |
Kpna6 |
karyopherin (importin) alpha 6 |
875 |
0.4 |
| chr1_36088323_36089060 | 0.99 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
12999 |
0.13 |
| chr5_136810958_136811326 | 0.92 |
Col26a1 |
collagen, type XXVI, alpha 1 |
27167 |
0.15 |
| chr1_189619014_189619289 | 0.92 |
Gm38122 |
predicted gene, 38122 |
21083 |
0.19 |
| chr19_53891604_53892011 | 0.91 |
Pdcd4 |
programmed cell death 4 |
424 |
0.79 |
| chr19_56378923_56379247 | 0.91 |
Nrap |
nebulin-related anchoring protein |
780 |
0.61 |
| chr18_20936839_20937401 | 0.90 |
Rnf125 |
ring finger protein 125 |
7505 |
0.22 |
| chr2_33335990_33336141 | 0.90 |
Gm13537 |
predicted gene 13537 |
12894 |
0.16 |
| chr10_80084809_80085161 | 0.86 |
Sbno2 |
strawberry notch 2 |
9546 |
0.09 |
| chr5_129179631_129179797 | 0.84 |
Rps16-ps2 |
ribosomal protein S16, pseudogene 2 |
51197 |
0.14 |
| chr2_30200927_30201078 | 0.83 |
Kyat1 |
kynurenine aminotransferase 1 |
2643 |
0.14 |
| chr4_142166267_142166486 | 0.83 |
Kazn |
kazrin, periplakin interacting protein |
7322 |
0.19 |
| chr3_84005748_84005921 | 0.82 |
Tmem131l |
transmembrane 131 like |
34294 |
0.18 |
| chr4_115822744_115822895 | 0.81 |
Mob3c |
MOB kinase activator 3C |
5273 |
0.13 |
| chr12_16544817_16544968 | 0.73 |
Lpin1 |
lipin 1 |
17146 |
0.24 |
| chr15_40674821_40674977 | 0.70 |
Zfpm2 |
zinc finger protein, multitype 2 |
9909 |
0.29 |
| chr17_12249866_12250017 | 0.70 |
Map3k4 |
mitogen-activated protein kinase kinase kinase 4 |
16045 |
0.14 |
| chr3_19635280_19635455 | 0.66 |
1700064H15Rik |
RIKEN cDNA 1700064H15 gene |
6690 |
0.16 |
| chr1_156987967_156988148 | 0.65 |
4930439D14Rik |
RIKEN cDNA 4930439D14 gene |
48219 |
0.1 |
| chr10_8912784_8913075 | 0.59 |
Gm48727 |
predicted gene, 48727 |
8948 |
0.16 |
| chr14_65718314_65718512 | 0.59 |
Scara5 |
scavenger receptor class A, member 5 |
41936 |
0.15 |
| chr12_64968157_64968335 | 0.56 |
Togaram1 |
TOG array regulator of axonemal microtubules 1 |
2190 |
0.21 |
| chr12_16731064_16731426 | 0.56 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
8883 |
0.17 |
| chr9_70044819_70045125 | 0.54 |
Gcnt3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
6884 |
0.14 |
| chr11_11928614_11928944 | 0.53 |
Grb10 |
growth factor receptor bound protein 10 |
9837 |
0.18 |
| chrX_60014607_60014758 | 0.53 |
F9 |
coagulation factor IX |
15218 |
0.19 |
| chr7_19160187_19160727 | 0.52 |
Gipr |
gastric inhibitory polypeptide receptor |
4637 |
0.08 |
| chr8_17053595_17053746 | 0.51 |
3110080E11Rik |
RIKEN cDNA 3110080E11 gene |
137488 |
0.05 |
| chr5_125203053_125203442 | 0.49 |
Ncor2 |
nuclear receptor co-repressor 2 |
24028 |
0.18 |
| chr1_92921240_92921447 | 0.46 |
Rnpepl1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
1729 |
0.19 |
| chr17_30006978_30007129 | 0.45 |
Zfand3 |
zinc finger, AN1-type domain 3 |
744 |
0.53 |
| chr1_85578306_85578542 | 0.44 |
G530012D18Rik |
RIKEN cDNA G530012D1 gene |
2748 |
0.14 |
| chr17_26497654_26497955 | 0.43 |
Gm34455 |
predicted gene, 34455 |
5396 |
0.12 |
| chr7_104507532_104507683 | 0.38 |
Trim30d |
tripartite motif-containing 30D |
218 |
0.86 |
| chr11_54250431_54250582 | 0.38 |
Csf2 |
colony stimulating factor 2 (granulocyte-macrophage) |
839 |
0.48 |
| chr18_74740901_74741052 | 0.37 |
Myo5b |
myosin VB |
7211 |
0.18 |
| chr4_85740566_85740717 | 0.35 |
Adamtsl1 |
ADAMTS-like 1 |
226314 |
0.02 |
| chr1_87397239_87397390 | 0.34 |
Kcnj13 |
potassium inwardly-rectifying channel, subfamily J, member 13 |
2585 |
0.2 |
| chr3_117001572_117002191 | 0.32 |
Gm9332 |
predicted gene 9332 |
2295 |
0.25 |
| chr4_133701094_133701537 | 0.32 |
Mir7227 |
microRNA 7227 |
15793 |
0.12 |
| chr4_131829199_131829486 | 0.29 |
Ptpru |
protein tyrosine phosphatase, receptor type, U |
7826 |
0.15 |
| chr16_34898145_34898335 | 0.27 |
Mylk |
myosin, light polypeptide kinase |
2015 |
0.36 |
| chr4_106827730_106828478 | 0.22 |
Gm12746 |
predicted gene 12746 |
19816 |
0.15 |
| chr1_40266407_40266915 | 0.22 |
Il1r1 |
interleukin 1 receptor, type I |
4 |
0.98 |
| chr2_146763416_146764171 | 0.22 |
Gm14111 |
predicted gene 14111 |
8088 |
0.27 |
| chr2_166080996_166081365 | 0.21 |
Sulf2 |
sulfatase 2 |
3061 |
0.24 |
| chr1_182053014_182053342 | 0.20 |
Gm37177 |
predicted gene, 37177 |
18225 |
0.14 |
| chr17_3436818_3437224 | 0.20 |
Tiam2 |
T cell lymphoma invasion and metastasis 2 |
2752 |
0.28 |
| chr7_128344992_128345271 | 0.19 |
Gm6916 |
predicted pseudogene 6916 |
4664 |
0.13 |
| chr7_92817657_92817808 | 0.17 |
Rab30 |
RAB30, member RAS oncogene family |
2173 |
0.27 |
| chr3_89696244_89696704 | 0.16 |
Adar |
adenosine deaminase, RNA-specific |
18548 |
0.14 |
| chr1_85116074_85116225 | 0.15 |
Gm16038 |
predicted gene 16038 |
830 |
0.42 |
| chr11_70970609_70971178 | 0.14 |
Rpain |
RPA interacting protein |
364 |
0.67 |
| chr2_6226731_6226967 | 0.13 |
Echdc3 |
enoyl Coenzyme A hydratase domain containing 3 |
13816 |
0.16 |
| chr14_21741906_21742090 | 0.13 |
Gm15935 |
predicted gene 15935 |
409 |
0.5 |
| chr8_126588694_126589418 | 0.12 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
4930 |
0.25 |
| chr8_10948388_10948539 | 0.12 |
Gm44955 |
predicted gene 44955 |
573 |
0.62 |
| chr5_64576943_64577119 | 0.12 |
Gm42566 |
predicted gene 42566 |
12212 |
0.12 |
| chr19_27228331_27228482 | 0.12 |
Vldlr |
very low density lipoprotein receptor |
10368 |
0.19 |
| chr8_128491772_128491970 | 0.09 |
Nrp1 |
neuropilin 1 |
132474 |
0.04 |
| chr5_15488572_15488741 | 0.09 |
Gm21149 |
predicted gene, 21149 |
12133 |
0.16 |
| chr10_86286801_86286952 | 0.07 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
13496 |
0.22 |
| chr8_26993871_26994644 | 0.05 |
Gm45370 |
predicted gene 45370 |
1993 |
0.18 |
| chr1_165091659_165092046 | 0.05 |
4930568G15Rik |
RIKEN cDNA 4930568G15 gene |
76 |
0.97 |
| chr1_133089589_133089740 | 0.04 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
21085 |
0.13 |
| chr3_144932528_144932679 | 0.04 |
Clca4b |
chloride channel accessory 4B |
74 |
0.96 |
| chr5_113804361_113804709 | 0.04 |
Tmem119 |
transmembrane protein 119 |
4019 |
0.12 |
| chr8_106054637_106054799 | 0.03 |
Nfatc3 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
4122 |
0.12 |
| chr5_25705272_25705934 | 0.03 |
Xrcc2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
161 |
0.93 |
| chr11_20460573_20460736 | 0.03 |
Gm12034 |
predicted gene 12034 |
14242 |
0.23 |
| chr9_110333949_110334114 | 0.01 |
Scap |
SREBF chaperone |
395 |
0.75 |
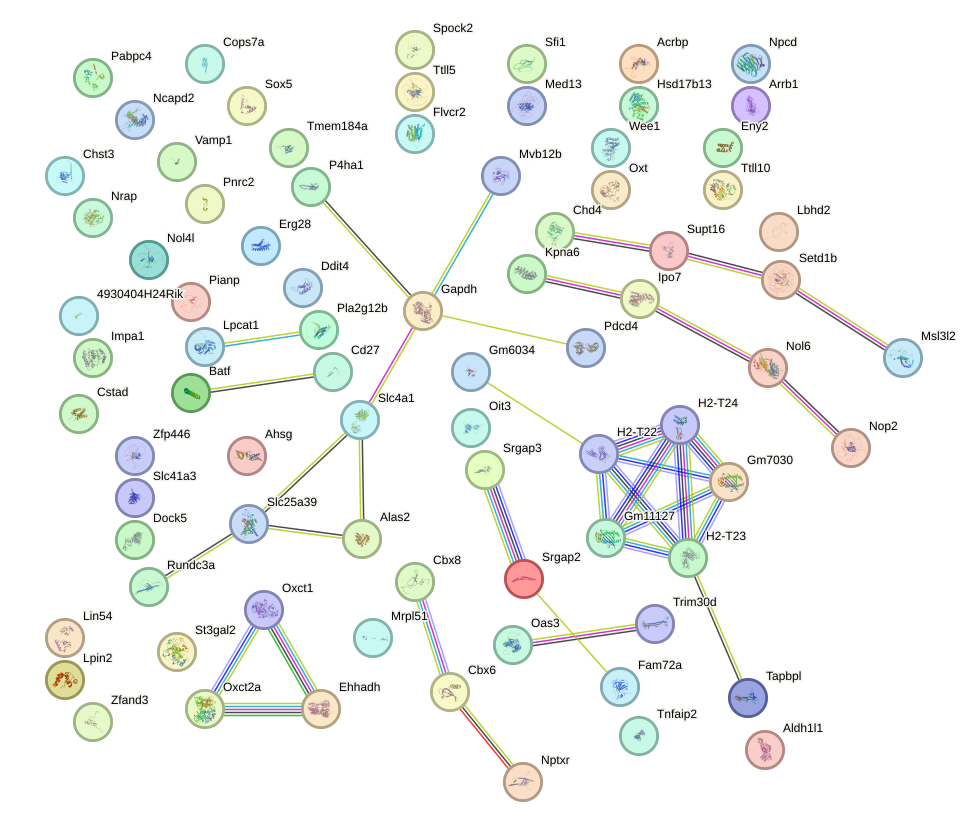
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 41.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.4 | 4.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 3.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.6 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.2 | 1.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 0.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 2.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 3.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.3 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.3 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.1 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.1 | 1.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.2 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 1.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.2 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.1 | 1.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 4.5 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.1 | 0.2 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.1 | 0.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 3.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.3 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 1.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) serotonin biosynthetic process(GO:0042427) neurotransmitter loading into synaptic vesicle(GO:0098700) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.9 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0045078 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.2 | GO:0048524 | positive regulation of viral process(GO:0048524) |
| 0.0 | 0.1 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.0 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.8 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of luteinizing hormone secretion(GO:0033685) negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 3.5 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) |
| 0.0 | 0.0 | GO:0034447 | glycoprotein transport(GO:0034436) very-low-density lipoprotein particle clearance(GO:0034447) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.0 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 41.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 4.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 1.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 3.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 3.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 41.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.8 | 3.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 4.4 | GO:0034778 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.3 | 3.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 1.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 1.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 1.5 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.4 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 3.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 3.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.3 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 1.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 1.4 | GO:0052830 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 1.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 2.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 42.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |