Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
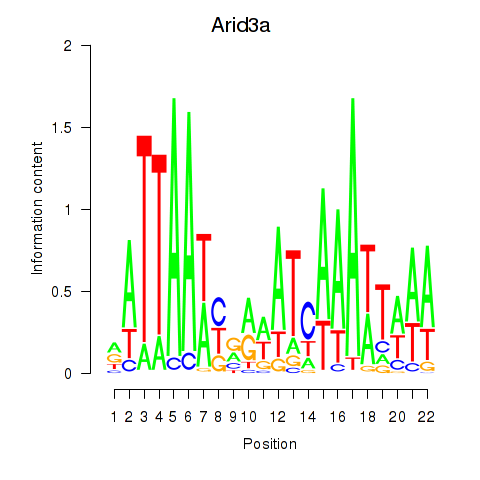
Results for Arid3a
Z-value: 1.10
Transcription factors associated with Arid3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid3a
|
ENSMUSG00000019564.6 | AT rich interactive domain 3A (BRIGHT-like) |
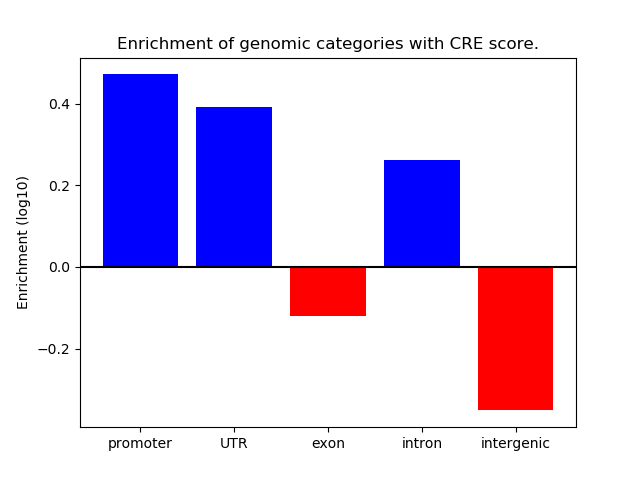
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_79937271_79937429 | Arid3a | 6926 | 0.061887 | 0.83 | 3.9e-02 | Click! |
| chr10_79926721_79927086 | Arid3a | 140 | 0.861880 | 0.80 | 5.6e-02 | Click! |
| chr10_79932814_79932968 | Arid3a | 2467 | 0.094376 | 0.76 | 8.1e-02 | Click! |
| chr10_79928248_79928404 | Arid3a | 985 | 0.244518 | 0.74 | 9.3e-02 | Click! |
| chr10_79928498_79928649 | Arid3a | 1232 | 0.186877 | 0.73 | 9.7e-02 | Click! |
Activity of the Arid3a motif across conditions
Conditions sorted by the z-value of the Arid3a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
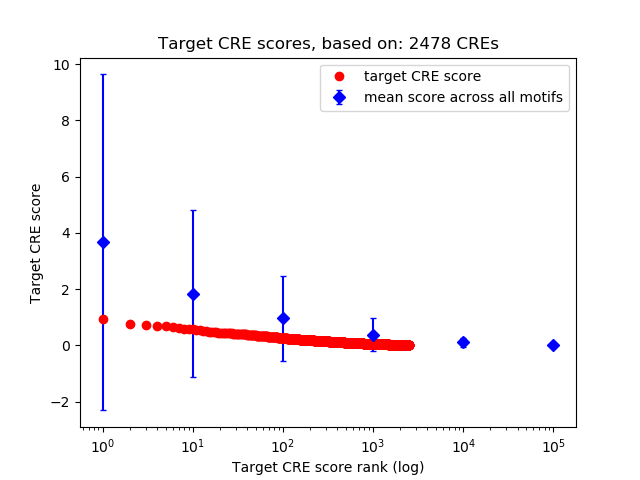
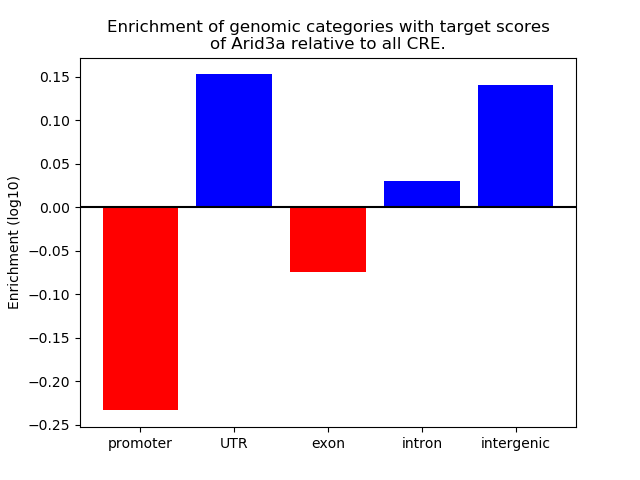
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_36537159_36537318 | 0.94 |
Ankrd23 |
ankyrin repeat domain 23 |
355 |
0.71 |
| chr10_31644816_31644968 | 0.75 |
Gm8793 |
predicted gene 8793 |
5734 |
0.23 |
| chr10_122384642_122385270 | 0.73 |
Gm36041 |
predicted gene, 36041 |
1936 |
0.39 |
| chr7_113223800_113224075 | 0.70 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
10686 |
0.21 |
| chr15_26353541_26353723 | 0.69 |
Marchf11 |
membrane associated ring-CH-type finger 11 |
34087 |
0.23 |
| chrX_11018346_11018497 | 0.67 |
Gm14485 |
predicted gene 14485 |
2586 |
0.39 |
| chrX_38473317_38473492 | 0.60 |
Gm7598 |
predicted gene 7598 |
3057 |
0.24 |
| chr5_8962552_8962757 | 0.60 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
4102 |
0.15 |
| chr18_20931708_20931938 | 0.57 |
Rnf125 |
ring finger protein 125 |
12802 |
0.2 |
| chr1_88760195_88760369 | 0.57 |
Platr5 |
pluripotency associated transcript 5 |
5328 |
0.21 |
| chr6_128504140_128504367 | 0.56 |
Pzp |
PZP, alpha-2-macroglobulin like |
9030 |
0.09 |
| chr18_20943771_20943971 | 0.56 |
Rnf125 |
ring finger protein 125 |
754 |
0.69 |
| chr14_100324093_100324244 | 0.53 |
Gm41231 |
predicted gene, 41231 |
38003 |
0.15 |
| chr6_24296762_24296913 | 0.49 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
128745 |
0.05 |
| chr2_154585868_154586214 | 0.49 |
E2f1 |
E2F transcription factor 1 |
16149 |
0.09 |
| chr6_128504617_128505108 | 0.48 |
Pzp |
PZP, alpha-2-macroglobulin like |
9639 |
0.09 |
| chr9_42253031_42253240 | 0.48 |
Sc5d |
sterol-C5-desaturase |
11065 |
0.17 |
| chr18_20921400_20921643 | 0.46 |
Rnf125 |
ring finger protein 125 |
23104 |
0.17 |
| chr5_134170212_134170373 | 0.45 |
Rcc1l |
reculator of chromosome condensation 1 like |
917 |
0.46 |
| chr5_135741186_135741337 | 0.45 |
Tmem120a |
transmembrane protein 120A |
1138 |
0.32 |
| chr15_56405270_56405431 | 0.45 |
Gm49213 |
predicted gene, 49213 |
9663 |
0.31 |
| chr11_8607022_8607173 | 0.45 |
Tns3 |
tensin 3 |
17790 |
0.28 |
| chr7_14430420_14431121 | 0.44 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
5677 |
0.17 |
| chr7_19866622_19866883 | 0.44 |
Gm44659 |
predicted gene 44659 |
4686 |
0.08 |
| chr9_77939601_77939775 | 0.43 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
1603 |
0.32 |
| chr13_10447671_10447864 | 0.43 |
Gm47407 |
predicted gene, 47407 |
69920 |
0.1 |
| chr8_124156032_124156199 | 0.43 |
Gm3889 |
predicted gene 3889 |
3958 |
0.23 |
| chr2_126549901_126550068 | 0.42 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
2423 |
0.3 |
| chr11_16704048_16704199 | 0.42 |
Gm25698 |
predicted gene, 25698 |
28588 |
0.15 |
| chr5_65621209_65621360 | 0.41 |
Pds5a |
PDS5 cohesin associated factor A |
684 |
0.49 |
| chr2_68873804_68874167 | 0.40 |
Cers6 |
ceramide synthase 6 |
12399 |
0.14 |
| chr4_118129721_118129877 | 0.40 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
5083 |
0.18 |
| chr5_144905134_144905285 | 0.40 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
7215 |
0.16 |
| chr16_42880363_42880514 | 0.40 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
4557 |
0.21 |
| chr5_23894319_23894470 | 0.39 |
AC117663.1 |
solute carrier family 40 (iron-regulated transporter), member 1 (Slc40a1), pseudogene |
17951 |
0.11 |
| chr5_150227882_150228033 | 0.39 |
Gm36378 |
predicted gene, 36378 |
15906 |
0.19 |
| chr2_85050098_85050267 | 0.39 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
278 |
0.85 |
| chr2_66712021_66712193 | 0.39 |
Scn7a |
sodium channel, voltage-gated, type VII, alpha |
72807 |
0.1 |
| chr10_68465783_68466111 | 0.39 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
59820 |
0.13 |
| chr12_109991265_109991416 | 0.38 |
Gm34667 |
predicted gene, 34667 |
32533 |
0.1 |
| chr13_93630762_93631092 | 0.38 |
Gm15622 |
predicted gene 15622 |
5545 |
0.17 |
| chr17_29056624_29056784 | 0.37 |
Gm41556 |
predicted gene, 41556 |
156 |
0.9 |
| chr3_83014473_83014624 | 0.37 |
Gm30097 |
predicted gene, 30097 |
6060 |
0.15 |
| chr16_29943850_29944001 | 0.37 |
Gm26569 |
predicted gene, 26569 |
2591 |
0.27 |
| chr12_35501929_35502080 | 0.37 |
Ahr |
aryl-hydrocarbon receptor |
24775 |
0.16 |
| chr3_104677278_104677438 | 0.37 |
Gm29560 |
predicted gene 29560 |
7348 |
0.1 |
| chr11_50642967_50643387 | 0.36 |
Gm12198 |
predicted gene 12198 |
40729 |
0.12 |
| chr12_62769518_62769809 | 0.36 |
Tspyl-ps |
testis-specific protein, Y-encoded-like, pseudogene |
65387 |
0.1 |
| chr5_33934831_33935145 | 0.36 |
Nelfa |
negative elongation factor complex member A, Whsc2 |
1262 |
0.31 |
| chr5_76043835_76044206 | 0.36 |
Gm32780 |
predicted gene, 32780 |
922 |
0.54 |
| chr4_11613136_11613395 | 0.34 |
Gm11832 |
predicted gene 11832 |
1742 |
0.3 |
| chr17_50397611_50397796 | 0.34 |
Gm49906 |
predicted gene, 49906 |
58451 |
0.13 |
| chr7_28832592_28832757 | 0.34 |
Lgals4 |
lectin, galactose binding, soluble 4 |
1180 |
0.23 |
| chr19_58413245_58413576 | 0.34 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
41056 |
0.17 |
| chr11_80430590_80430899 | 0.34 |
Psmd11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
857 |
0.6 |
| chr14_116308331_116308711 | 0.34 |
Gm20713 |
predicted gene 20713 |
437243 |
0.01 |
| chr17_93740961_93741187 | 0.34 |
Gm50003 |
predicted gene, 50003 |
60040 |
0.14 |
| chr16_28841105_28841256 | 0.33 |
Mb21d2 |
Mab-21 domain containing 2 |
4813 |
0.32 |
| chr1_151327603_151327859 | 0.33 |
Gm10138 |
predicted gene 10138 |
16351 |
0.14 |
| chr12_21142640_21142894 | 0.33 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30813 |
0.16 |
| chr2_144078197_144078361 | 0.33 |
Banf2os |
barrier to autointegration factor 2, opposite strand |
9473 |
0.18 |
| chr7_84106850_84107019 | 0.33 |
Cemip |
cell migration inducing protein, hyaluronan binding |
20432 |
0.16 |
| chr9_110326292_110326443 | 0.33 |
Gm42638 |
predicted gene 42638 |
4197 |
0.13 |
| chr1_59425292_59425875 | 0.33 |
Gm37737 |
predicted gene, 37737 |
23919 |
0.15 |
| chr13_37667494_37667917 | 0.32 |
AI463229 |
expressed sequence AI463229 |
29 |
0.96 |
| chr4_142077888_142078052 | 0.31 |
Tmem51os1 |
Tmem51 opposite strand 1 |
6002 |
0.15 |
| chr16_91448666_91448817 | 0.31 |
Gm46562 |
predicted gene, 46562 |
9680 |
0.1 |
| chr2_126729966_126730117 | 0.31 |
Gm10774 |
predicted pseudogene 10774 |
20484 |
0.13 |
| chr19_46899287_46899584 | 0.31 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
10140 |
0.16 |
| chr11_48771756_48771907 | 0.31 |
Gm12183 |
predicted gene 12183 |
18117 |
0.09 |
| chr7_19928412_19928563 | 0.31 |
Pvr |
poliovirus receptor |
7327 |
0.08 |
| chr1_186827890_186828061 | 0.30 |
A430105J06Rik |
RIKEN cDNA A430105J06 gene |
78579 |
0.07 |
| chr6_147064387_147064568 | 0.30 |
Mrps35 |
mitochondrial ribosomal protein S35 |
3226 |
0.17 |
| chr13_52972702_52972854 | 0.30 |
n-R5s54 |
nuclear encoded rRNA 5S 54 |
8046 |
0.18 |
| chr15_31063560_31063731 | 0.30 |
4930430F21Rik |
RIKEN cDNA 4930430F21 gene |
24517 |
0.19 |
| chr8_128463488_128463809 | 0.29 |
Nrp1 |
neuropilin 1 |
104251 |
0.07 |
| chr1_139548791_139549059 | 0.29 |
Cfhr1 |
complement factor H-related 1 |
4674 |
0.22 |
| chr17_73839522_73839982 | 0.29 |
Gm4948 |
predicted gene 4948 |
31598 |
0.14 |
| chr10_61310200_61310351 | 0.29 |
Rpl27a-ps1 |
ribosomal protein L27A, pseudogene 1 |
445 |
0.76 |
| chr10_28556799_28556950 | 0.28 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
3277 |
0.37 |
| chr7_123193136_123193287 | 0.28 |
Tnrc6a |
trinucleotide repeat containing 6a |
13290 |
0.2 |
| chr5_8899522_8899888 | 0.28 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
5756 |
0.16 |
| chr16_17727505_17727656 | 0.28 |
Gm6048 |
predicted gene 6048 |
2204 |
0.14 |
| chr14_75179701_75179900 | 0.28 |
Lcp1 |
lymphocyte cytosolic protein 1 |
3592 |
0.19 |
| chr8_14825690_14825955 | 0.28 |
Dlgap2 |
DLG associated protein 2 |
48049 |
0.12 |
| chr12_106479388_106479572 | 0.28 |
Gm3191 |
predicted gene 3191 |
20330 |
0.16 |
| chr2_155539663_155540189 | 0.28 |
Mipep-ps |
mitochondrial intermediate peptidase, pseudogene |
2721 |
0.13 |
| chr10_8089089_8089240 | 0.28 |
Gm48614 |
predicted gene, 48614 |
67872 |
0.11 |
| chr16_14177645_14177827 | 0.28 |
Nde1 |
nudE neurodevelopment protein 1 |
2609 |
0.22 |
| chr19_56602273_56602424 | 0.28 |
Nhlrc2 |
NHL repeat containing 2 |
31551 |
0.15 |
| chr2_157550429_157550705 | 0.27 |
Gm14286 |
predicted gene 14286 |
5098 |
0.12 |
| chr8_71405347_71405498 | 0.27 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
588 |
0.54 |
| chr19_20757249_20757433 | 0.27 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
29779 |
0.2 |
| chr14_20319722_20320198 | 0.26 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr19_25405865_25406016 | 0.26 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
759 |
0.7 |
| chr4_35013042_35013212 | 0.26 |
Gm12364 |
predicted gene 12364 |
50907 |
0.12 |
| chr19_30128744_30128991 | 0.26 |
Gldc |
glycine decarboxylase |
16364 |
0.18 |
| chr9_70270024_70270175 | 0.26 |
Myo1e |
myosin IE |
62731 |
0.11 |
| chr7_75467233_75467387 | 0.26 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
11442 |
0.16 |
| chr4_19681355_19681795 | 0.25 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
3127 |
0.29 |
| chr4_99178772_99179088 | 0.25 |
Atg4c |
autophagy related 4C, cysteine peptidase |
15004 |
0.16 |
| chr1_139660727_139661253 | 0.25 |
Cfhr3 |
complement factor H-related 3 |
91 |
0.97 |
| chrX_12004590_12004741 | 0.25 |
Gm14512 |
predicted gene 14512 |
20197 |
0.22 |
| chr2_33444795_33444946 | 0.25 |
Gm13536 |
predicted gene 13536 |
1932 |
0.27 |
| chr10_61486909_61487060 | 0.25 |
Gm47595 |
predicted gene, 47595 |
9306 |
0.1 |
| chr16_24394028_24394581 | 0.25 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr2_61401897_61402378 | 0.25 |
Gm22338 |
predicted gene, 22338 |
90629 |
0.09 |
| chr18_20976494_20976645 | 0.25 |
Rnf125 |
ring finger protein 125 |
15097 |
0.19 |
| chr3_21935040_21935222 | 0.24 |
Gm43674 |
predicted gene 43674 |
63337 |
0.12 |
| chr1_93693606_93693762 | 0.24 |
Bok |
BCL2-related ovarian killer |
133 |
0.94 |
| chr15_89210708_89210873 | 0.24 |
Ppp6r2 |
protein phosphatase 6, regulatory subunit 2 |
763 |
0.48 |
| chr1_191254917_191255072 | 0.24 |
Gm37074 |
predicted gene, 37074 |
5570 |
0.14 |
| chr7_113778913_113779066 | 0.24 |
Spon1 |
spondin 1, (f-spondin) extracellular matrix protein |
12815 |
0.22 |
| chr10_80621807_80621958 | 0.24 |
Csnk1g2 |
casein kinase 1, gamma 2 |
956 |
0.3 |
| chr5_32195986_32196216 | 0.24 |
Gm9555 |
predicted gene 9555 |
10604 |
0.15 |
| chr19_29268750_29269067 | 0.24 |
Jak2 |
Janus kinase 2 |
16467 |
0.17 |
| chr13_64235436_64235605 | 0.24 |
Cdc14b |
CDC14 cell division cycle 14B |
13122 |
0.1 |
| chr11_17501097_17501506 | 0.24 |
Gm12016 |
predicted gene 12016 |
137882 |
0.05 |
| chr1_106177771_106177975 | 0.24 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
6121 |
0.19 |
| chr14_41170829_41171191 | 0.23 |
Sftpd |
surfactant associated protein D |
12125 |
0.11 |
| chr2_103293475_103293769 | 0.23 |
Ehf |
ets homologous factor |
2685 |
0.27 |
| chr12_86478264_86478415 | 0.23 |
Esrrb |
estrogen related receptor, beta |
8222 |
0.26 |
| chr12_34268416_34268623 | 0.23 |
Gm18025 |
predicted gene, 18025 |
22616 |
0.27 |
| chr9_36795863_36796248 | 0.23 |
Ei24 |
etoposide induced 2.4 mRNA |
965 |
0.44 |
| chr2_122781620_122782092 | 0.23 |
Sqor |
sulfide quinone oxidoreductase |
143 |
0.96 |
| chr10_128272971_128273122 | 0.23 |
Stat2 |
signal transducer and activator of transcription 2 |
2470 |
0.11 |
| chr3_83012044_83012325 | 0.23 |
Gm30097 |
predicted gene, 30097 |
3696 |
0.17 |
| chr1_190204546_190204725 | 0.23 |
Prox1 |
prospero homeobox 1 |
33921 |
0.16 |
| chr3_28736802_28736989 | 0.23 |
1700112D23Rik |
RIKEN cDNA 1700112D23 gene |
5345 |
0.16 |
| chr12_101739231_101739640 | 0.22 |
Tc2n |
tandem C2 domains, nuclear |
20912 |
0.2 |
| chr2_103625976_103626181 | 0.22 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
59768 |
0.1 |
| chr16_37606843_37607024 | 0.22 |
Gm46559 |
predicted gene, 46559 |
10743 |
0.15 |
| chr6_116910457_116910623 | 0.22 |
Gm43926 |
predicted gene, 43926 |
42979 |
0.16 |
| chr13_114457557_114457872 | 0.22 |
Fst |
follistatin |
872 |
0.39 |
| chr5_34296898_34297150 | 0.22 |
Cfap99 |
cilia and flagella associated protein 99 |
8424 |
0.11 |
| chr2_165921904_165922055 | 0.22 |
Gm11461 |
predicted gene 11461 |
3862 |
0.16 |
| chr1_78649448_78649613 | 0.22 |
Acsl3 |
acyl-CoA synthetase long-chain family member 3 |
8295 |
0.16 |
| chr2_173047391_173047542 | 0.22 |
Gm14453 |
predicted gene 14453 |
12886 |
0.13 |
| chr15_67469080_67469242 | 0.22 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
242392 |
0.02 |
| chr2_114776001_114776450 | 0.21 |
Gm13975 |
predicted gene 13975 |
68113 |
0.11 |
| chr12_8217539_8217881 | 0.21 |
Gm33037 |
predicted gene, 33037 |
8545 |
0.19 |
| chr13_37512858_37513176 | 0.21 |
Gm29590 |
predicted gene 29590 |
3299 |
0.14 |
| chr19_23024753_23025259 | 0.21 |
Gm50136 |
predicted gene, 50136 |
36448 |
0.17 |
| chr10_83324249_83324444 | 0.21 |
Slc41a2 |
solute carrier family 41, member 2 |
1463 |
0.37 |
| chr8_48545544_48545713 | 0.21 |
Tenm3 |
teneurin transmembrane protein 3 |
9685 |
0.29 |
| chr9_69987908_69988201 | 0.21 |
Bnip2 |
BCL2/adenovirus E1B interacting protein 2 |
1412 |
0.32 |
| chr4_132570302_132570458 | 0.21 |
Ptafr |
platelet-activating factor receptor |
6313 |
0.12 |
| chr13_59771624_59772060 | 0.21 |
Etohd2 |
ethanol decreased 2 |
1876 |
0.15 |
| chr12_85818362_85818513 | 0.21 |
Erg28 |
ergosterol biosynthesis 28 |
3813 |
0.2 |
| chr10_20616725_20616876 | 0.21 |
Gm17230 |
predicted gene 17230 |
8835 |
0.25 |
| chr13_28860566_28860730 | 0.21 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
22808 |
0.15 |
| chr18_38431542_38431707 | 0.21 |
Ndfip1 |
Nedd4 family interacting protein 1 |
12640 |
0.15 |
| chr6_128515921_128516189 | 0.21 |
Pzp |
PZP, alpha-2-macroglobulin like |
10648 |
0.09 |
| chr19_33099137_33099698 | 0.20 |
Gm29946 |
predicted gene, 29946 |
23230 |
0.18 |
| chr1_78650012_78650396 | 0.20 |
Acsl3 |
acyl-CoA synthetase long-chain family member 3 |
7621 |
0.17 |
| chr2_60947339_60947519 | 0.20 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
15763 |
0.22 |
| chr2_6718585_6718742 | 0.20 |
Celf2 |
CUGBP, Elav-like family member 2 |
2950 |
0.38 |
| chr5_87135400_87135653 | 0.20 |
Ugt2b5 |
UDP glucuronosyltransferase 2 family, polypeptide B5 |
4792 |
0.14 |
| chr17_15341574_15341736 | 0.20 |
Gm7423 |
predicted gene 7423 |
19773 |
0.14 |
| chr2_130578101_130578388 | 0.20 |
Oxt |
oxytocin |
2071 |
0.16 |
| chr5_125405640_125405791 | 0.20 |
Dhx37 |
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
10182 |
0.11 |
| chr2_130961670_130961943 | 0.20 |
Atrn |
attractin |
20979 |
0.12 |
| chr7_132931708_132932726 | 0.20 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
1020 |
0.34 |
| chr2_165015545_165015729 | 0.20 |
Ncoa5 |
nuclear receptor coactivator 5 |
7576 |
0.14 |
| chr5_18002991_18003251 | 0.20 |
Gnat3 |
guanine nucleotide binding protein, alpha transducing 3 |
40572 |
0.21 |
| chr11_86963788_86963969 | 0.20 |
Ypel2 |
yippee like 2 |
8146 |
0.19 |
| chr11_110011350_110011662 | 0.20 |
Abca8b |
ATP-binding cassette, sub-family A (ABC1), member 8b |
15661 |
0.2 |
| chr8_86639656_86639807 | 0.20 |
Lonp2 |
lon peptidase 2, peroxisomal |
8334 |
0.17 |
| chr5_87335535_87335718 | 0.19 |
Ugt2a3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
1569 |
0.26 |
| chr2_115592873_115593046 | 0.19 |
BC052040 |
cDNA sequence BC052040 |
11158 |
0.19 |
| chr14_66080264_66080431 | 0.19 |
Adam2 |
a disintegrin and metallopeptidase domain 2 |
2614 |
0.24 |
| chr5_140204413_140204753 | 0.19 |
Gm16120 |
predicted gene 16120 |
2802 |
0.25 |
| chr19_41829163_41829397 | 0.19 |
Frat1 |
frequently rearranged in advanced T cell lymphomas |
690 |
0.62 |
| chr11_80146295_80146446 | 0.19 |
Tefm |
transcription elongation factor, mitochondrial |
4195 |
0.16 |
| chr5_135776480_135776870 | 0.19 |
Styxl1 |
serine/threonine/tyrosine interacting-like 1 |
1563 |
0.22 |
| chr10_44698108_44698268 | 0.19 |
Gm47388 |
predicted gene, 47388 |
1008 |
0.41 |
| chr14_59548598_59548749 | 0.19 |
Cdadc1 |
cytidine and dCMP deaminase domain containing 1 |
49163 |
0.11 |
| chr5_41572654_41572805 | 0.19 |
Rab28 |
RAB28, member RAS oncogene family |
54773 |
0.16 |
| chr13_51841406_51841559 | 0.19 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
5196 |
0.25 |
| chr17_29381584_29381926 | 0.19 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
5254 |
0.14 |
| chr1_179836199_179836367 | 0.19 |
Ahctf1 |
AT hook containing transcription factor 1 |
32603 |
0.16 |
| chr12_106479176_106479338 | 0.19 |
Gm3191 |
predicted gene 3191 |
20107 |
0.16 |
| chr2_132654710_132655013 | 0.19 |
Rpl23a-ps4 |
ribosomal protein L23A, pseudogene 4 |
7381 |
0.12 |
| chr5_8902050_8902207 | 0.19 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
8179 |
0.15 |
| chr1_84877752_84878205 | 0.19 |
Fbxo36 |
F-box protein 36 |
8827 |
0.16 |
| chr11_70746635_70746805 | 0.19 |
Gm12320 |
predicted gene 12320 |
8431 |
0.08 |
| chr6_91514715_91514905 | 0.19 |
Xpc |
xeroderma pigmentosum, complementation group C |
985 |
0.32 |
| chr19_33097519_33097677 | 0.19 |
Gm29946 |
predicted gene, 29946 |
25049 |
0.18 |
| chr5_126565776_126566015 | 0.19 |
Gm24839 |
predicted gene, 24839 |
51914 |
0.14 |
| chr9_55474509_55474664 | 0.19 |
Etfa |
electron transferring flavoprotein, alpha polypeptide |
12927 |
0.19 |
| chr1_61649997_61650160 | 0.19 |
Gm37205 |
predicted gene, 37205 |
4345 |
0.17 |
| chr4_134930449_134930699 | 0.19 |
Syf2 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
324 |
0.86 |
| chr6_51540654_51541195 | 0.19 |
Snx10 |
sorting nexin 10 |
3599 |
0.26 |
| chr7_15086187_15086338 | 0.19 |
Gtpbp4-ps4 |
GTP binding protein 4, pseudogene 4 |
764 |
0.45 |
| chr10_25563128_25563279 | 0.19 |
Gm29571 |
predicted gene 29571 |
26817 |
0.14 |
| chr17_86458349_86458750 | 0.18 |
Prkce |
protein kinase C, epsilon |
32013 |
0.19 |
| chr3_108640818_108640982 | 0.18 |
Clcc1 |
chloride channel CLIC-like 1 |
13013 |
0.11 |
| chr2_103451907_103452157 | 0.18 |
Elf5 |
E74-like factor 5 |
3294 |
0.23 |
| chr6_30563747_30563898 | 0.18 |
Cpa4 |
carboxypeptidase A4 |
4547 |
0.14 |
| chr17_84269883_84270034 | 0.18 |
Gm24492 |
predicted gene, 24492 |
17789 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.2 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.0 | GO:1900193 | regulation of oocyte maturation(GO:1900193) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |