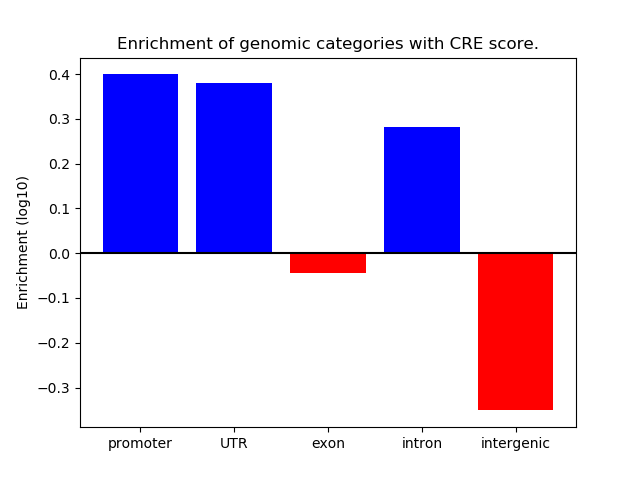
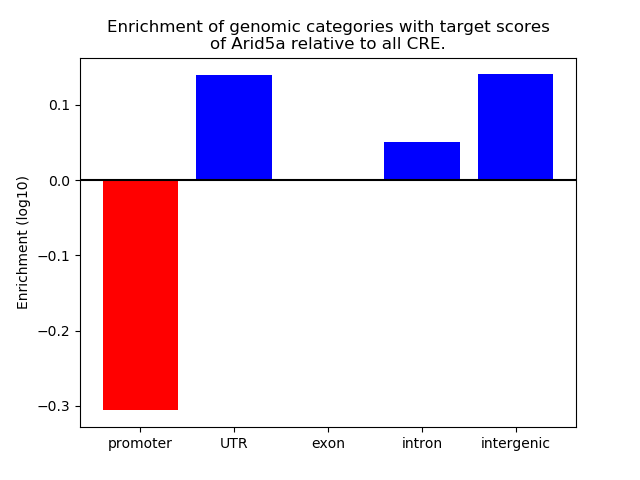
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
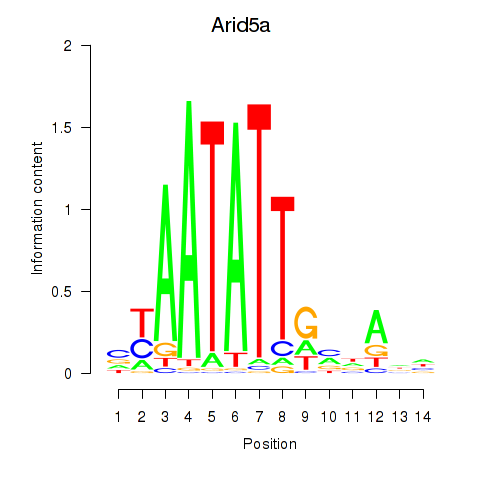
Results for Arid5a
Z-value: 1.33
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSMUSG00000037447.10 | AT rich interactive domain 5A (MRF1-like) |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_36303522_36303898 | Arid5a | 4023 | 0.162436 | 0.76 | 8.1e-02 | Click! |
| chr1_36324139_36324297 | Arid5a | 7191 | 0.129238 | -0.68 | 1.4e-01 | Click! |
| chr1_36307583_36307861 | Arid5a | 11 | 0.967711 | 0.51 | 3.0e-01 | Click! |
| chr1_36307915_36308291 | Arid5a | 324 | 0.844588 | -0.21 | 6.9e-01 | Click! |
| chr1_36308411_36308603 | Arid5a | 728 | 0.577620 | -0.10 | 8.5e-01 | Click! |
Activity of the Arid5a motif across conditions
Conditions sorted by the z-value of the Arid5a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_110290286_110290470 | 1.19 |
Bbox1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
15363 |
0.21 |
| chr7_112620610_112620794 | 0.74 |
Gm24154 |
predicted gene, 24154 |
2477 |
0.27 |
| chr10_122361947_122362104 | 0.64 |
Gm36041 |
predicted gene, 36041 |
24867 |
0.2 |
| chr5_145852474_145852695 | 0.61 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
24107 |
0.13 |
| chr11_41470910_41471082 | 0.55 |
Hspd1-ps3 |
heat shock protein 1 (chaperonin), pseudogene 3 |
27741 |
0.23 |
| chr14_102790892_102791046 | 0.54 |
Gm23764 |
predicted gene, 23764 |
15130 |
0.23 |
| chr6_128482999_128483184 | 0.50 |
Pzp |
PZP, alpha-2-macroglobulin like |
4764 |
0.1 |
| chr10_19585787_19585961 | 0.50 |
Ifngr1 |
interferon gamma receptor 1 |
6075 |
0.2 |
| chr3_57386543_57386727 | 0.40 |
Gm5276 |
predicted gene 5276 |
24247 |
0.18 |
| chr5_77406298_77406449 | 0.36 |
Igfbp7 |
insulin-like growth factor binding protein 7 |
1667 |
0.3 |
| chr11_7610322_7610520 | 0.35 |
Gm27393 |
predicted gene, 27393 |
276084 |
0.01 |
| chr14_51106134_51106584 | 0.34 |
Eddm3b |
epididymal protein 3B |
8067 |
0.08 |
| chr4_123530925_123531089 | 0.34 |
Macf1 |
microtubule-actin crosslinking factor 1 |
3359 |
0.27 |
| chr8_89207397_89207570 | 0.33 |
Gm5356 |
predicted pseudogene 5356 |
19923 |
0.24 |
| chr15_99836517_99836668 | 0.33 |
Lima1 |
LIM domain and actin binding 1 |
16449 |
0.08 |
| chr14_57345054_57345267 | 0.32 |
Gm23593 |
predicted gene, 23593 |
11675 |
0.15 |
| chr7_99899121_99899281 | 0.32 |
Xrra1 |
X-ray radiation resistance associated 1 |
3505 |
0.15 |
| chr13_40857803_40857954 | 0.32 |
Gcnt2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
1876 |
0.2 |
| chr15_27560020_27560329 | 0.32 |
Ank |
progressive ankylosis |
11273 |
0.16 |
| chr4_126091997_126092189 | 0.31 |
Oscp1 |
organic solute carrier partner 1 |
4414 |
0.13 |
| chr10_108979346_108979586 | 0.31 |
Gm47477 |
predicted gene, 47477 |
22978 |
0.22 |
| chr10_63410268_63410432 | 0.31 |
Gm7530 |
predicted gene 7530 |
2649 |
0.19 |
| chr9_100619599_100619756 | 0.31 |
Gm8661 |
predicted gene 8661 |
6288 |
0.15 |
| chr1_23282801_23282987 | 0.30 |
Gm27028 |
predicted gene, 27028 |
8643 |
0.12 |
| chr15_83544542_83544693 | 0.28 |
Bik |
BCL2-interacting killer |
2079 |
0.19 |
| chr14_117680704_117680890 | 0.28 |
Mir6239 |
microRNA 6239 |
273050 |
0.01 |
| chr15_41842516_41842689 | 0.27 |
Oxr1 |
oxidation resistance 1 |
7622 |
0.21 |
| chr5_142426503_142426820 | 0.27 |
Foxk1 |
forkhead box K1 |
25161 |
0.17 |
| chr3_148845994_148846145 | 0.26 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
10544 |
0.29 |
| chr10_20589812_20590002 | 0.26 |
Gm17230 |
predicted gene 17230 |
35728 |
0.16 |
| chr19_54071291_54071462 | 0.26 |
Gm50186 |
predicted gene, 50186 |
13183 |
0.18 |
| chr8_108731775_108731954 | 0.25 |
Gm38042 |
predicted gene, 38042 |
5729 |
0.25 |
| chr12_84938982_84939198 | 0.25 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
4288 |
0.14 |
| chr1_192808464_192808629 | 0.25 |
Gm38360 |
predicted gene, 38360 |
12581 |
0.13 |
| chr8_45264437_45264634 | 0.25 |
F11 |
coagulation factor XI |
2504 |
0.25 |
| chr13_69263863_69264031 | 0.24 |
Gm4812 |
predicted gene 4812 |
81708 |
0.09 |
| chr19_32269255_32269433 | 0.23 |
Sgms1 |
sphingomyelin synthase 1 |
7582 |
0.25 |
| chr14_50990821_50990973 | 0.23 |
Gm49038 |
predicted gene, 49038 |
6178 |
0.08 |
| chr18_20945531_20945682 | 0.23 |
Rnf125 |
ring finger protein 125 |
981 |
0.59 |
| chr5_102856822_102856973 | 0.23 |
Arhgap24 |
Rho GTPase activating protein 24 |
11890 |
0.28 |
| chr12_43274258_43274412 | 0.23 |
Gm47030 |
predicted gene, 47030 |
31808 |
0.25 |
| chr5_20256525_20256681 | 0.23 |
Gm43680 |
predicted gene 43680 |
4359 |
0.23 |
| chr13_52902494_52902653 | 0.23 |
Auh |
AU RNA binding protein/enoyl-coenzyme A hydratase |
10373 |
0.24 |
| chr17_64741176_64741339 | 0.23 |
Gm17133 |
predicted gene 17133 |
12839 |
0.19 |
| chr14_36969125_36969474 | 0.23 |
Ccser2 |
coiled-coil serine rich 2 |
522 |
0.8 |
| chr2_159725785_159725995 | 0.22 |
Gm11445 |
predicted gene 11445 |
52317 |
0.17 |
| chr9_86648283_86648497 | 0.22 |
Me1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
29653 |
0.11 |
| chr17_73839991_73840145 | 0.22 |
Gm4948 |
predicted gene 4948 |
31282 |
0.15 |
| chr7_102281548_102281715 | 0.22 |
Stim1 |
stromal interaction molecule 1 |
13304 |
0.13 |
| chr8_64790589_64790740 | 0.22 |
Klhl2 |
kelch-like 2, Mayven |
9000 |
0.16 |
| chr5_25529146_25529503 | 0.22 |
1700096K18Rik |
RIKEN cDNA 1700096K18 gene |
689 |
0.57 |
| chr10_52802004_52802190 | 0.22 |
Gm47623 |
predicted gene, 47623 |
604 |
0.76 |
| chr2_65226961_65227145 | 0.21 |
Cobll1 |
Cobl-like 1 |
8942 |
0.19 |
| chr14_41167667_41167818 | 0.21 |
Sftpd |
surfactant associated protein D |
15393 |
0.1 |
| chr18_45588547_45588708 | 0.21 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
14007 |
0.23 |
| chrX_143532306_143532457 | 0.21 |
Pak3 |
p21 (RAC1) activated kinase 3 |
13683 |
0.28 |
| chr10_24086637_24086815 | 0.20 |
Taar8b |
trace amine-associated receptor 8B |
5568 |
0.09 |
| chrX_12377036_12377378 | 0.20 |
Gm14635 |
predicted gene 14635 |
22211 |
0.24 |
| chr1_180757526_180757875 | 0.20 |
Gm37768 |
predicted gene, 37768 |
1864 |
0.22 |
| chr1_15817639_15817823 | 0.20 |
Terf1 |
telomeric repeat binding factor 1 |
10090 |
0.18 |
| chr4_147084058_147084372 | 0.20 |
Gm13160 |
predicted gene 13160 |
2769 |
0.16 |
| chr10_25331794_25331983 | 0.20 |
Akap7 |
A kinase (PRKA) anchor protein 7 |
24018 |
0.15 |
| chr12_84198545_84198912 | 0.19 |
Gm31513 |
predicted gene, 31513 |
2759 |
0.15 |
| chrX_93589200_93589367 | 0.19 |
Gm23557 |
predicted gene, 23557 |
15415 |
0.18 |
| chr6_99145207_99145385 | 0.19 |
Foxp1 |
forkhead box P1 |
17722 |
0.26 |
| chr19_26600604_26600755 | 0.19 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
4371 |
0.21 |
| chr18_46630413_46630795 | 0.19 |
Gm3734 |
predicted gene 3734 |
203 |
0.92 |
| chr2_104702604_104702799 | 0.19 |
Tcp11l1 |
t-complex 11 like 1 |
3766 |
0.18 |
| chr15_16252844_16253026 | 0.19 |
Gm6479 |
predicted gene 6479 |
44982 |
0.2 |
| chr12_40610701_40610983 | 0.19 |
Dock4 |
dedicator of cytokinesis 4 |
164506 |
0.03 |
| chr11_99739497_99739661 | 0.19 |
Gm14180 |
predicted gene 14180 |
5289 |
0.06 |
| chr15_7878227_7878378 | 0.18 |
Wdr70 |
WD repeat domain 70 |
8075 |
0.23 |
| chr8_123813297_123813472 | 0.18 |
Rab4a |
RAB4A, member RAS oncogene family |
7316 |
0.1 |
| chr13_100025978_100026249 | 0.18 |
Mccc2 |
methylcrotonoyl-Coenzyme A carboxylase 2 (beta) |
10474 |
0.19 |
| chr17_84702223_84702399 | 0.18 |
Abcg8 |
ATP binding cassette subfamily G member 8 |
19180 |
0.13 |
| chr13_100533300_100533570 | 0.18 |
Ocln |
occludin |
19025 |
0.11 |
| chr4_19942325_19942476 | 0.17 |
4930480G23Rik |
RIKEN cDNA 4930480G23 gene |
538 |
0.8 |
| chr2_77518455_77518668 | 0.17 |
Zfp385b |
zinc finger protein 385B |
974 |
0.66 |
| chr7_30921191_30921414 | 0.17 |
Hamp2 |
hepcidin antimicrobial peptide 2 |
2879 |
0.09 |
| chr3_157956049_157956385 | 0.17 |
Ankrd13c |
ankyrin repeat domain 13c |
8751 |
0.13 |
| chr13_96746399_96746550 | 0.17 |
Ankrd31 |
ankyrin repeat domain 31 |
1798 |
0.34 |
| chr6_88717537_88717719 | 0.17 |
Mgll |
monoglyceride lipase |
6784 |
0.13 |
| chr6_143472792_143472951 | 0.17 |
Gm23272 |
predicted gene, 23272 |
7474 |
0.28 |
| chr4_129928314_129928470 | 0.17 |
Spocd1 |
SPOC domain containing 1 |
857 |
0.48 |
| chr5_84169095_84169246 | 0.17 |
Hmgn2-ps1 |
high mobility group nucleosomal binding domain 2, pseudogene 1 |
45857 |
0.2 |
| chr2_154609874_154610025 | 0.17 |
Zfp341 |
zinc finger protein 341 |
3348 |
0.13 |
| chr10_21259230_21259397 | 0.17 |
Gm33728 |
predicted gene, 33728 |
7199 |
0.16 |
| chr5_63955081_63955232 | 0.17 |
Rell1 |
RELT-like 1 |
13667 |
0.15 |
| chr5_105826898_105827072 | 0.16 |
Lrrc8d |
leucine rich repeat containing 8D |
2474 |
0.21 |
| chr10_40016495_40016677 | 0.16 |
Gm8055 |
predicted pseudogene 8055 |
4720 |
0.13 |
| chr9_43453811_43453984 | 0.16 |
Gm47475 |
predicted gene, 47475 |
9459 |
0.18 |
| chr4_116462908_116463073 | 0.16 |
Mast2 |
microtubule associated serine/threonine kinase 2 |
761 |
0.6 |
| chr2_140159319_140159521 | 0.16 |
Esf1 |
ESF1 nucleolar pre-rRNA processing protein homolog |
577 |
0.72 |
| chr6_36280777_36280955 | 0.16 |
9330158H04Rik |
RIKEN cDNA 9330158H04 gene |
6510 |
0.29 |
| chr8_10912890_10913092 | 0.16 |
Gm2814 |
predicted gene 2814 |
12433 |
0.09 |
| chr11_50175754_50175938 | 0.16 |
Mrnip |
MRN complex interacting protein |
939 |
0.42 |
| chr2_65229254_65229444 | 0.16 |
Cobll1 |
Cobl-like 1 |
6646 |
0.2 |
| chr7_46855536_46855687 | 0.16 |
Ldhc |
lactate dehydrogenase C |
5592 |
0.1 |
| chr5_139084403_139084554 | 0.16 |
Prkar1b |
protein kinase, cAMP dependent regulatory, type I beta |
28166 |
0.15 |
| chr13_41596510_41596690 | 0.16 |
Tmem170b |
transmembrane protein 170B |
9307 |
0.16 |
| chr5_144335608_144335778 | 0.16 |
Dmrt1i |
Dmrt1 interacting ncRNA |
22230 |
0.14 |
| chr2_49452901_49453060 | 0.16 |
Epc2 |
enhancer of polycomb homolog 2 |
1494 |
0.5 |
| chr7_30191689_30191840 | 0.16 |
Capns1 |
calpain, small subunit 1 |
646 |
0.42 |
| chr3_98677782_98678103 | 0.16 |
Gm12399 |
predicted gene 12399 |
389 |
0.8 |
| chr1_9798859_9799037 | 0.15 |
Sgk3 |
serum/glucocorticoid regulated kinase 3 |
740 |
0.56 |
| chr11_83078008_83078159 | 0.15 |
Gm20234 |
predicted gene, 20234 |
6837 |
0.08 |
| chr13_10445581_10445750 | 0.15 |
Gm47407 |
predicted gene, 47407 |
72022 |
0.1 |
| chr15_96375345_96375521 | 0.15 |
Arid2 |
AT rich interactive domain 2 (ARID, RFX-like) |
3381 |
0.25 |
| chr5_73539093_73539252 | 0.15 |
Dcun1d4 |
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
28304 |
0.15 |
| chr10_22153823_22154174 | 0.15 |
Gm4895 |
predicted gene 4895 |
418 |
0.7 |
| chr10_85183539_85183728 | 0.15 |
Cry1 |
cryptochrome 1 (photolyase-like) |
1431 |
0.44 |
| chrX_16846156_16846327 | 0.15 |
Maob |
monoamine oxidase B |
28875 |
0.23 |
| chr6_5449067_5449234 | 0.15 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
47111 |
0.15 |
| chr2_4444586_4444844 | 0.15 |
Gm13175 |
predicted gene 13175 |
21955 |
0.17 |
| chr16_36526263_36526432 | 0.15 |
Casr |
calcium-sensing receptor |
35259 |
0.09 |
| chr11_100330647_100330968 | 0.15 |
Gast |
gastrin |
3600 |
0.1 |
| chr9_56553589_56553770 | 0.14 |
Gm47178 |
predicted gene, 47178 |
9923 |
0.16 |
| chr6_82039277_82039610 | 0.14 |
Eva1a |
eva-1 homolog A (C. elegans) |
1600 |
0.34 |
| chr9_64235160_64235318 | 0.14 |
Uchl4 |
ubiquitin carboxyl-terminal esterase L4 |
38 |
0.95 |
| chr2_119546518_119546691 | 0.14 |
Exd1 |
exonuclease 3'-5' domain containing 1 |
590 |
0.55 |
| chr8_35077237_35077419 | 0.14 |
Gm34419 |
predicted gene, 34419 |
17217 |
0.16 |
| chr3_34482441_34482598 | 0.14 |
Gm29135 |
predicted gene 29135 |
312 |
0.9 |
| chr14_21576402_21576583 | 0.14 |
Kat6b |
K(lysine) acetyltransferase 6B |
46050 |
0.15 |
| chrX_57712526_57712689 | 0.14 |
Gm8107 |
predicted gene 8107 |
25948 |
0.14 |
| chr18_64009153_64009310 | 0.14 |
Gm6974 |
predicted gene 6974 |
72189 |
0.09 |
| chr6_35874026_35874908 | 0.14 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr8_71603005_71603180 | 0.14 |
Fam129c |
family with sequence similarity 129, member C |
741 |
0.42 |
| chr19_38356216_38356572 | 0.14 |
Gm50150 |
predicted gene, 50150 |
13780 |
0.14 |
| chr8_23190059_23190210 | 0.14 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
804 |
0.48 |
| chr1_105671022_105671199 | 0.14 |
Relch |
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
6669 |
0.16 |
| chr11_108251538_108251704 | 0.14 |
Gm11655 |
predicted gene 11655 |
69771 |
0.1 |
| chr19_23062879_23063069 | 0.14 |
Gm50136 |
predicted gene, 50136 |
1501 |
0.41 |
| chr2_118451174_118451332 | 0.14 |
Eif2ak4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
5907 |
0.17 |
| chr8_29633674_29633854 | 0.14 |
Nudc-ps1 |
nuclear distribution gene C homolog (Aspergillus), pseudogene 1 |
346783 |
0.01 |
| chr13_37667494_37667917 | 0.14 |
AI463229 |
expressed sequence AI463229 |
29 |
0.96 |
| chr8_93161284_93161435 | 0.14 |
Gm45910 |
predicted gene 45910 |
7243 |
0.14 |
| chr7_132610799_132611117 | 0.14 |
Lhpp |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
250 |
0.9 |
| chr17_75714219_75714387 | 0.14 |
Gm9360 |
predicted gene 9360 |
53063 |
0.16 |
| chr4_97007725_97007914 | 0.13 |
Gm27521 |
predicted gene, 27521 |
90799 |
0.09 |
| chr8_128481354_128481549 | 0.13 |
Nrp1 |
neuropilin 1 |
122054 |
0.05 |
| chr4_54954102_54954261 | 0.13 |
Zfp462 |
zinc finger protein 462 |
6205 |
0.29 |
| chr7_45471496_45471656 | 0.13 |
Bax |
BCL2-associated X protein |
4678 |
0.06 |
| chr9_77948137_77948457 | 0.13 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
7006 |
0.15 |
| chr10_63373741_63373935 | 0.13 |
Sirt1 |
sirtuin 1 |
7866 |
0.13 |
| chr1_39068328_39068494 | 0.13 |
Gm37821 |
predicted gene, 37821 |
10774 |
0.18 |
| chr18_46644335_46644497 | 0.13 |
Gm3734 |
predicted gene 3734 |
13609 |
0.15 |
| chr11_77659536_77659706 | 0.13 |
Gm23132 |
predicted gene, 23132 |
914 |
0.46 |
| chr12_37906736_37906912 | 0.13 |
Dgkb |
diacylglycerol kinase, beta |
26108 |
0.21 |
| chr4_144896444_144896602 | 0.13 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
3304 |
0.27 |
| chr8_116404702_116404855 | 0.13 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
47717 |
0.17 |
| chr13_33956943_33957100 | 0.13 |
Nqo2 |
N-ribosyldihydronicotinamide quinone reductase 2 |
7666 |
0.13 |
| chr14_54579168_54579536 | 0.13 |
Ajuba |
ajuba LIM protein |
1794 |
0.15 |
| chrX_101233242_101233419 | 0.13 |
Gm14842 |
predicted gene 14842 |
4663 |
0.11 |
| chr6_113302370_113302539 | 0.13 |
Gm43935 |
predicted gene, 43935 |
4220 |
0.1 |
| chr3_129585968_129586158 | 0.13 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
33683 |
0.13 |
| chr2_107625700_107625874 | 0.13 |
Gm9864 |
predicted gene 9864 |
25248 |
0.27 |
| chr8_45672138_45672498 | 0.13 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
10322 |
0.21 |
| chr14_21232114_21232287 | 0.13 |
Adk |
adenosine kinase |
85925 |
0.09 |
| chr10_68435331_68435497 | 0.13 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
90353 |
0.08 |
| chr4_109842744_109842984 | 0.13 |
Faf1 |
Fas-associated factor 1 |
2079 |
0.41 |
| chr10_33812363_33812541 | 0.13 |
Gm10327 |
predicted pseudogene 10327 |
2954 |
0.21 |
| chr11_35280936_35281100 | 0.13 |
Gm12123 |
predicted gene 12123 |
42652 |
0.16 |
| chr2_108847431_108847673 | 0.13 |
Rpl35a-ps4 |
ribosomal protein 35A, pseudogene 4 |
27706 |
0.23 |
| chr10_21444614_21444790 | 0.13 |
Gm48386 |
predicted gene, 48386 |
934 |
0.49 |
| chr2_43673499_43673650 | 0.13 |
Kynu |
kynureninase |
2644 |
0.4 |
| chr3_121904528_121904954 | 0.13 |
Gm42593 |
predicted gene 42593 |
41899 |
0.12 |
| chr2_124555012_124555366 | 0.12 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
54917 |
0.17 |
| chr4_116647902_116648068 | 0.12 |
Akr1a1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
1489 |
0.26 |
| chr3_85265616_85265908 | 0.12 |
1700036G14Rik |
RIKEN cDNA 1700036G14 gene |
51757 |
0.14 |
| chr7_90026148_90026300 | 0.12 |
E230029C05Rik |
RIKEN cDNA E230029C05 gene |
2935 |
0.18 |
| chr7_110942759_110942910 | 0.12 |
Mrvi1 |
MRV integration site 1 |
3353 |
0.24 |
| chr4_14993690_14993896 | 0.12 |
Gm11844 |
predicted gene 11844 |
28789 |
0.2 |
| chr5_23386690_23386916 | 0.12 |
4930580E04Rik |
RIKEN cDNA 4930580E04 gene |
306 |
0.8 |
| chr1_102734086_102734402 | 0.12 |
Gm22034 |
predicted gene, 22034 |
219304 |
0.02 |
| chr15_83223287_83223580 | 0.12 |
A4galt |
alpha 1,4-galactosyltransferase |
28296 |
0.1 |
| chr4_145066440_145066605 | 0.12 |
Vps13d |
vacuolar protein sorting 13D |
6080 |
0.27 |
| chr15_75986268_75986587 | 0.12 |
Ccdc166 |
coiled-coil domain containing 166 |
3972 |
0.09 |
| chr9_58591565_58591716 | 0.12 |
Nptn |
neuroplastin |
9196 |
0.18 |
| chr9_32242875_32243026 | 0.12 |
Arhgap32 |
Rho GTPase activating protein 32 |
2214 |
0.34 |
| chr11_31101231_31101397 | 0.12 |
Gm12105 |
predicted gene 12105 |
18689 |
0.18 |
| chr11_97440527_97441283 | 0.12 |
Arhgap23 |
Rho GTPase activating protein 23 |
4620 |
0.18 |
| chr15_59046761_59047065 | 0.12 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6316 |
0.23 |
| chr10_40259071_40259222 | 0.12 |
Rnu3a |
U3A small nuclear RNA |
679 |
0.41 |
| chr14_25527411_25527588 | 0.12 |
Mir3075 |
microRNA 3075 |
6940 |
0.17 |
| chr5_65611228_65611632 | 0.12 |
Pds5a |
PDS5 cohesin associated factor A |
343 |
0.77 |
| chr8_111808698_111808849 | 0.11 |
Cfdp1 |
craniofacial development protein 1 |
45518 |
0.11 |
| chr18_8933795_8934011 | 0.11 |
Gm37148 |
predicted gene, 37148 |
5668 |
0.27 |
| chr10_54049674_54049878 | 0.11 |
Gm47917 |
predicted gene, 47917 |
14035 |
0.19 |
| chr5_146828447_146828685 | 0.11 |
Rpl21 |
ribosomal protein L21 |
4324 |
0.14 |
| chr9_65776050_65776201 | 0.11 |
Zfp609 |
zinc finger protein 609 |
21787 |
0.15 |
| chr8_11747362_11747524 | 0.11 |
Gm18991 |
predicted gene, 18991 |
8822 |
0.11 |
| chr8_108706681_108707155 | 0.11 |
Zfhx3 |
zinc finger homeobox 3 |
3818 |
0.29 |
| chr15_44856570_44856766 | 0.11 |
A930017M01Rik |
RIKEN cDNA A930017M01 gene |
24751 |
0.19 |
| chr13_28528509_28528789 | 0.11 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
7541 |
0.23 |
| chr4_40725671_40725834 | 0.11 |
Dnaja1 |
DnaJ heat shock protein family (Hsp40) member A1 |
1703 |
0.23 |
| chr17_88604336_88604567 | 0.11 |
Gm9406 |
predicted gene 9406 |
4093 |
0.19 |
| chr5_147017691_147017860 | 0.11 |
Lnx2 |
ligand of numb-protein X 2 |
1260 |
0.45 |
| chr7_140856325_140856484 | 0.11 |
Gm45785 |
predicted gene 45785 |
73 |
0.48 |
| chr4_86847350_86847506 | 0.11 |
Rps6 |
ribosomal protein S6 |
9904 |
0.19 |
| chr2_103714634_103714812 | 0.11 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
312 |
0.88 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |