Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
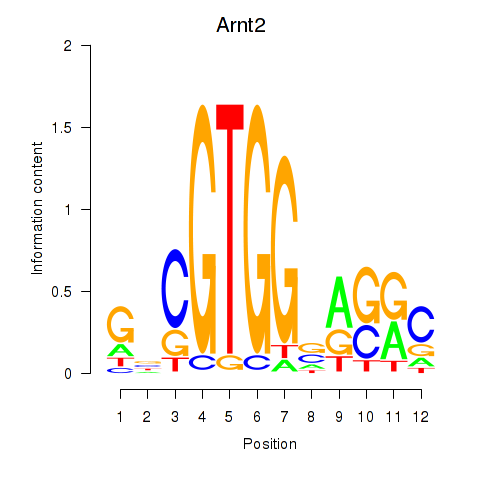
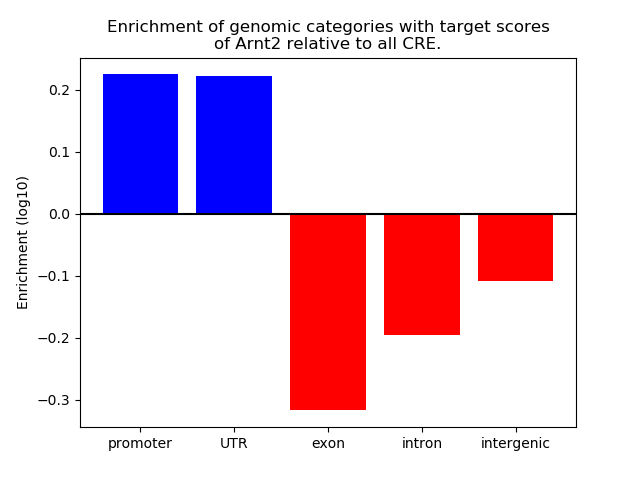
Results for Arnt2
Z-value: 0.30
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.8 | aryl hydrocarbon receptor nuclear translocator 2 |
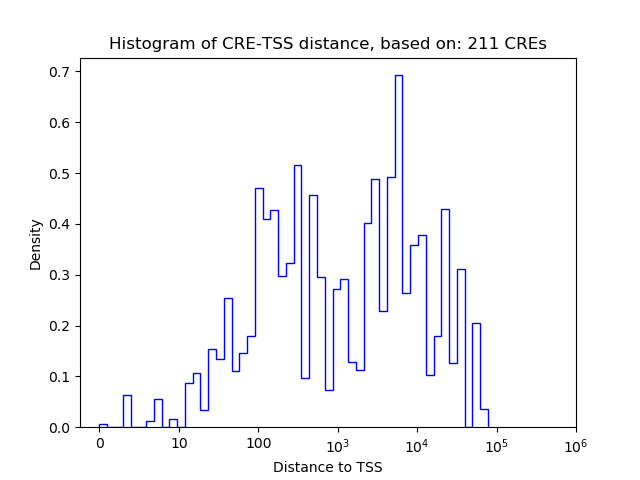
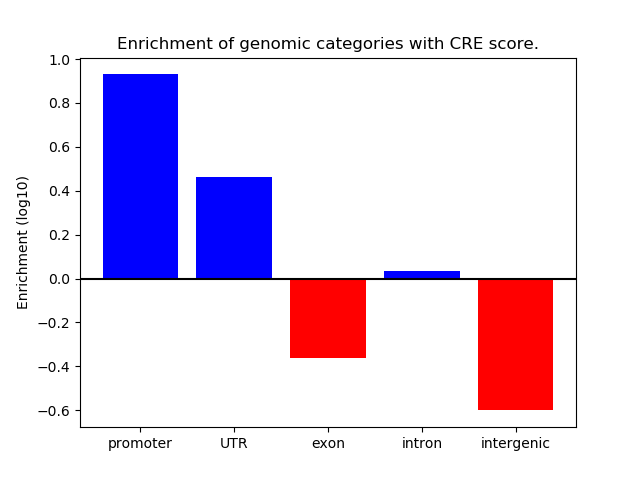
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_84355719_84355915 | Arnt2 | 12094 | 0.159467 | -0.89 | 1.8e-02 | Click! |
| chr7_84409303_84409461 | Arnt2 | 501 | 0.775351 | 0.76 | 7.9e-02 | Click! |
| chr7_84272524_84272705 | Arnt2 | 3257 | 0.219422 | 0.54 | 2.7e-01 | Click! |
| chr7_84417366_84417553 | Arnt2 | 7283 | 0.169754 | -0.51 | 3.1e-01 | Click! |
| chr7_84417589_84417755 | Arnt2 | 7496 | 0.168838 | -0.45 | 3.7e-01 | Click! |
Activity of the Arnt2 motif across conditions
Conditions sorted by the z-value of the Arnt2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
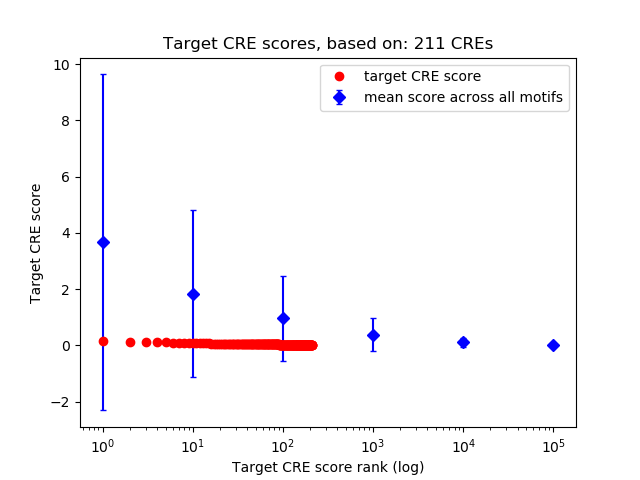
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_28898892_28899474 | 0.17 |
Gm31508 |
predicted gene, 31508 |
11046 |
0.17 |
| chr3_83040161_83040458 | 0.11 |
Fgb |
fibrinogen beta chain |
9554 |
0.14 |
| chr12_3858258_3858439 | 0.11 |
Dnmt3a |
DNA methyltransferase 3A |
2673 |
0.24 |
| chr11_70608766_70608939 | 0.11 |
Mink1 |
misshapen-like kinase 1 (zebrafish) |
886 |
0.3 |
| chr5_118560059_118560217 | 0.10 |
Med13l |
mediator complex subunit 13-like |
541 |
0.72 |
| chr9_107908957_107909108 | 0.10 |
Mst1r |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
2125 |
0.13 |
| chr9_78022288_78022639 | 0.09 |
Gm47829 |
predicted gene, 47829 |
5482 |
0.12 |
| chr7_107600772_107600937 | 0.09 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
5569 |
0.16 |
| chr1_71938041_71938229 | 0.08 |
Gm28818 |
predicted gene 28818 |
21010 |
0.17 |
| chr7_128398804_128398955 | 0.07 |
Rgs10 |
regulator of G-protein signalling 10 |
5194 |
0.13 |
| chr9_70243258_70243430 | 0.07 |
Myo1e |
myosin IE |
35976 |
0.17 |
| chr9_48480379_48480530 | 0.07 |
Rexo2 |
RNA exonuclease 2 |
125 |
0.95 |
| chr1_106759665_106759986 | 0.07 |
Kdsr |
3-ketodihydrosphingosine reductase |
98 |
0.97 |
| chr16_32065993_32066164 | 0.07 |
Gm26419 |
predicted gene, 26419 |
168 |
0.91 |
| chr14_25660817_25660989 | 0.07 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
4073 |
0.19 |
| chr12_111816647_111816859 | 0.06 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
497 |
0.67 |
| chr8_106621324_106621497 | 0.06 |
Cdh1 |
cadherin 1 |
17267 |
0.16 |
| chr8_123234508_123234799 | 0.06 |
4933417D19Rik |
RIKEN cDNA 4933417D19 gene |
438 |
0.57 |
| chr17_25472162_25472329 | 0.06 |
Tekt4 |
tektin 4 |
648 |
0.56 |
| chr8_124751425_124751592 | 0.06 |
Fam89a |
family with sequence similarity 89, member A |
324 |
0.83 |
| chr13_58359003_58359169 | 0.06 |
Kif27 |
kinesin family member 27 |
36 |
0.96 |
| chr15_36960858_36961018 | 0.06 |
Gm34590 |
predicted gene, 34590 |
22074 |
0.14 |
| chr4_142077888_142078052 | 0.06 |
Tmem51os1 |
Tmem51 opposite strand 1 |
6002 |
0.15 |
| chr2_33130281_33130432 | 0.05 |
Garnl3 |
GTPase activating RANGAP domain-like 3 |
252 |
0.91 |
| chr7_31115327_31115506 | 0.05 |
Hpn |
hepsin |
126 |
0.92 |
| chr19_8920279_8920446 | 0.05 |
B3gat3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
12 |
0.91 |
| chr3_21988216_21988409 | 0.05 |
Gm43674 |
predicted gene 43674 |
10156 |
0.21 |
| chr11_44405632_44405787 | 0.05 |
Il12b |
interleukin 12b |
5646 |
0.2 |
| chr5_140648568_140648783 | 0.05 |
Ttyh3 |
tweety family member 3 |
322 |
0.85 |
| chr9_107655632_107656026 | 0.05 |
Slc38a3 |
solute carrier family 38, member 3 |
191 |
0.86 |
| chr17_84877556_84877740 | 0.05 |
Gm49982 |
predicted gene, 49982 |
24921 |
0.14 |
| chr11_101083529_101083687 | 0.05 |
Coasy |
Coenzyme A synthase |
287 |
0.78 |
| chr11_50292381_50292544 | 0.05 |
Maml1 |
mastermind like transcriptional coactivator 1 |
151 |
0.93 |
| chr10_128459073_128459224 | 0.05 |
Smarcc2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
100 |
0.81 |
| chr5_88929617_88929827 | 0.05 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
4810 |
0.28 |
| chr13_43252482_43252652 | 0.05 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
50838 |
0.13 |
| chr10_79866111_79866298 | 0.05 |
Plppr3 |
phospholipid phosphatase related 3 |
1077 |
0.19 |
| chr12_86810853_86811037 | 0.04 |
Gm10095 |
predicted gene 10095 |
35522 |
0.14 |
| chr5_88961784_88961935 | 0.04 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
27287 |
0.22 |
| chr6_120777539_120777793 | 0.04 |
Slc25a18 |
solute carrier family 25 (mitochondrial carrier), member 18 |
4088 |
0.17 |
| chr11_59787617_59787784 | 0.04 |
Pld6 |
phospholipase D family, member 6 |
55 |
0.95 |
| chr1_191575029_191575351 | 0.04 |
Dtl |
denticleless E3 ubiquitin protein ligase |
321 |
0.58 |
| chr8_72496145_72496313 | 0.04 |
Gm17435 |
predicted gene, 17435 |
3285 |
0.11 |
| chr7_130865692_130865852 | 0.04 |
Plekha1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
16 |
0.91 |
| chr6_88831379_88831579 | 0.04 |
Abtb1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
7236 |
0.11 |
| chr3_121263297_121263469 | 0.04 |
Tlcd4 |
TLC domain containing 4 |
43 |
0.97 |
| chr13_56588544_56588882 | 0.04 |
2010203P06Rik |
RIKEN cDNA 2010203P06 gene |
6824 |
0.18 |
| chr6_39205376_39205554 | 0.04 |
Kdm7a |
lysine (K)-specific demethylase 7A |
1324 |
0.32 |
| chr16_85172553_85172704 | 0.04 |
App |
amyloid beta (A4) precursor protein |
1076 |
0.5 |
| chr19_6855220_6855477 | 0.04 |
Ccdc88b |
coiled-coil domain containing 88B |
110 |
0.93 |
| chr5_66080941_66081107 | 0.04 |
Rbm47 |
RNA binding motif protein 47 |
24 |
0.96 |
| chr14_63370757_63370955 | 0.04 |
Blk |
B lymphoid kinase |
5562 |
0.18 |
| chr17_56673508_56673661 | 0.04 |
Ranbp3 |
RAN binding protein 3 |
200 |
0.9 |
| chr9_120539742_120539893 | 0.04 |
Entpd3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
1 |
0.96 |
| chr7_35146051_35146625 | 0.04 |
Gm35665 |
predicted gene, 35665 |
4889 |
0.12 |
| chr2_124764238_124764402 | 0.04 |
Gm13994 |
predicted gene 13994 |
55665 |
0.16 |
| chr7_127703919_127704239 | 0.04 |
Bcl7c |
B cell CLL/lymphoma 7C |
3225 |
0.1 |
| chr7_30072560_30072736 | 0.04 |
Zfp82 |
zinc finger protein 82 |
153 |
0.89 |
| chr10_80702547_80702707 | 0.04 |
Izumo4 |
IZUMO family member 4 |
97 |
0.87 |
| chr4_133680298_133680626 | 0.04 |
Pigv |
phosphatidylinositol glycan anchor biosynthesis, class V |
7815 |
0.13 |
| chr7_49527548_49528235 | 0.04 |
Nav2 |
neuron navigator 2 |
20301 |
0.23 |
| chr8_70839238_70839453 | 0.04 |
Arrdc2 |
arrestin domain containing 2 |
173 |
0.87 |
| chr15_76207439_76207714 | 0.04 |
Plec |
plectin |
677 |
0.46 |
| chr9_109092894_109093071 | 0.04 |
Plxnb1 |
plexin B1 |
2407 |
0.14 |
| chr5_140607202_140607368 | 0.04 |
Lfng |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
35 |
0.93 |
| chr10_81192883_81193034 | 0.04 |
Dapk3 |
death-associated protein kinase 3 |
1402 |
0.16 |
| chr4_11147830_11148134 | 0.04 |
Gm11830 |
predicted gene 11830 |
3272 |
0.15 |
| chr11_84682675_84682945 | 0.03 |
1700109G15Rik |
RIKEN cDNA 1700109G15 gene |
37622 |
0.16 |
| chr3_138742123_138742276 | 0.03 |
Tspan5 |
tetraspanin 5 |
4 |
0.98 |
| chr16_17132359_17132527 | 0.03 |
Sdf2l1 |
stromal cell-derived factor 2-like 1 |
60 |
0.92 |
| chr17_46328441_46328616 | 0.03 |
Abcc10 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
176 |
0.92 |
| chr8_120438150_120438320 | 0.03 |
Gm22715 |
predicted gene, 22715 |
5314 |
0.21 |
| chr11_78826748_78827038 | 0.03 |
Lyrm9 |
LYR motif containing 9 |
264 |
0.89 |
| chr4_141723409_141723567 | 0.03 |
Ddi2 |
DNA-damage inducible protein 2 |
69 |
0.96 |
| chr5_53998780_53998931 | 0.03 |
Stim2 |
stromal interaction molecule 2 |
290 |
0.93 |
| chr6_82652519_82652678 | 0.03 |
Pole4 |
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
78 |
0.97 |
| chr16_95814486_95814819 | 0.03 |
2810404F17Rik |
RIKEN cDNA 2810404F17 gene |
6335 |
0.19 |
| chr1_171247148_171247313 | 0.03 |
Ndufs2 |
NADH:ubiquinone oxidoreductase core subunit S2 |
108 |
0.9 |
| chr1_191313013_191313425 | 0.03 |
Nenf |
neuron derived neurotrophic factor |
4975 |
0.16 |
| chr3_122482421_122482595 | 0.03 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
1215 |
0.39 |
| chr2_27212341_27212510 | 0.03 |
Sardh |
sarcosine dehydrogenase |
2488 |
0.21 |
| chr9_60837448_60837607 | 0.03 |
Gm9869 |
predicted gene 9869 |
672 |
0.71 |
| chr3_60057962_60058174 | 0.03 |
Sucnr1 |
succinate receptor 1 |
23801 |
0.14 |
| chr4_117229573_117229739 | 0.03 |
1700012C08Rik |
RIKEN cDNA 1700012C08 gene |
15874 |
0.07 |
| chr2_154552446_154552618 | 0.03 |
Actl10 |
actin-like 10 |
756 |
0.49 |
| chr7_118491860_118492044 | 0.03 |
Itpripl2 |
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
23 |
0.97 |
| chr8_94377700_94377868 | 0.03 |
Gm15889 |
predicted gene 15889 |
88 |
0.94 |
| chr4_155992309_155992490 | 0.03 |
B3galt6 |
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6 |
250 |
0.63 |
| chr8_84712564_84712722 | 0.03 |
Nfix |
nuclear factor I/X |
1425 |
0.25 |
| chr8_107478885_107479169 | 0.03 |
Wwp2 |
WW domain containing E3 ubiquitin protein ligase 2 |
4305 |
0.19 |
| chr17_26508062_26508231 | 0.03 |
Dusp1 |
dual specificity phosphatase 1 |
326 |
0.81 |
| chrX_98937926_98938238 | 0.03 |
Yipf6 |
Yip1 domain family, member 6 |
125 |
0.97 |
| chr10_127516737_127516940 | 0.03 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
1871 |
0.17 |
| chr8_11730699_11730857 | 0.03 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
2601 |
0.15 |
| chr4_139964484_139964679 | 0.03 |
Mir2139 |
microRNA 2139 |
3039 |
0.2 |
| chr9_44537727_44537883 | 0.03 |
Cxcr5 |
chemokine (C-X-C motif) receptor 5 |
11223 |
0.08 |
| chr1_86595445_86595606 | 0.03 |
Cops7b |
COP9 signalosome subunit 7B |
1215 |
0.35 |
| chr17_55712577_55712746 | 0.03 |
Pot1b |
protection of telomeres 1B |
33 |
0.97 |
| chr19_5567813_5567988 | 0.03 |
Ap5b1 |
adaptor-related protein complex 5, beta 1 subunit |
125 |
0.9 |
| chr14_11569795_11569971 | 0.03 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
16302 |
0.21 |
| chr5_43233554_43233922 | 0.03 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
189 |
0.9 |
| chr10_127349953_127350109 | 0.02 |
Inhbe |
inhibin beta-E |
4380 |
0.09 |
| chr11_82567439_82567600 | 0.02 |
Gm24612 |
predicted gene, 24612 |
28660 |
0.17 |
| chr11_75196330_75196483 | 0.02 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
2623 |
0.15 |
| chr5_142819812_142819971 | 0.02 |
Tnrc18 |
trinucleotide repeat containing 18 |
2229 |
0.28 |
| chr8_47673422_47673581 | 0.02 |
Ing2 |
inhibitor of growth family, member 2 |
188 |
0.9 |
| chr5_65536628_65536812 | 0.02 |
Smim14 |
small integral membrane protein 14 |
409 |
0.45 |
| chr1_36094936_36095089 | 0.02 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
6678 |
0.14 |
| chr1_88213972_88214137 | 0.02 |
Ugt1a1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 |
2095 |
0.13 |
| chr11_69684271_69684422 | 0.02 |
Tnfsf13os |
tumor necrosis factor (ligand) superfamily, member 13, opposite strand |
17 |
0.89 |
| chr9_90260181_90260562 | 0.02 |
Tbc1d2b |
TBC1 domain family, member 2B |
4444 |
0.2 |
| chr4_141139422_141139603 | 0.02 |
Szrd1 |
SUZ RNA binding domain containing 1 |
215 |
0.89 |
| chr8_126986591_126986750 | 0.02 |
Gm38574 |
predicted gene, 38574 |
4438 |
0.17 |
| chr7_28919694_28920029 | 0.02 |
Actn4 |
actinin alpha 4 |
640 |
0.55 |
| chrX_167719562_167719742 | 0.02 |
Gm15238 |
predicted gene 15238 |
56609 |
0.15 |
| chr12_80103737_80103943 | 0.02 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
9154 |
0.12 |
| chr4_118138797_118138948 | 0.02 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
3958 |
0.19 |
| chr17_5189712_5190024 | 0.02 |
Gm15599 |
predicted gene 15599 |
77758 |
0.1 |
| chr11_57697009_57697179 | 0.02 |
4933426K07Rik |
RIKEN cDNA 4933426K07 gene |
39268 |
0.11 |
| chr11_43747505_43747851 | 0.02 |
Ttc1 |
tetratricopeptide repeat domain 1 |
304 |
0.91 |
| chr8_69654338_69654500 | 0.02 |
Zfp964 |
zinc finger protein 964 |
60 |
0.97 |
| chr11_74896549_74896739 | 0.02 |
Sgsm2 |
small G protein signaling modulator 2 |
416 |
0.66 |
| chr11_69125215_69125587 | 0.02 |
Aloxe3 |
arachidonate lipoxygenase 3 |
495 |
0.58 |
| chr4_130662896_130663297 | 0.02 |
Pum1 |
pumilio RNA-binding family member 1 |
225 |
0.94 |
| chr7_16098491_16098656 | 0.02 |
Napa |
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
35 |
0.96 |
| chr5_32139274_32139441 | 0.02 |
Fosl2 |
fos-like antigen 2 |
3184 |
0.2 |
| chr9_18473377_18473557 | 0.02 |
Zfp558 |
zinc finger protein 558 |
45 |
0.96 |
| chr1_82291509_82291798 | 0.02 |
Irs1 |
insulin receptor substrate 1 |
237 |
0.93 |
| chr7_111082364_111082563 | 0.02 |
Eif4g2 |
eukaryotic translation initiation factor 4, gamma 2 |
268 |
0.88 |
| chr2_32353075_32353226 | 0.02 |
Dnm1 |
dynamin 1 |
84 |
0.9 |
| chr19_6401728_6401968 | 0.02 |
Rasgrp2 |
RAS, guanyl releasing protein 2 |
129 |
0.91 |
| chr2_27625052_27625210 | 0.02 |
Rxra |
retinoid X receptor alpha |
51309 |
0.12 |
| chr15_11407473_11407634 | 0.02 |
Tars |
threonyl-tRNA synthetase |
7888 |
0.27 |
| chr9_44510210_44510366 | 0.02 |
Bcl9l |
B cell CLL/lymphoma 9-like |
11016 |
0.07 |
| chr17_15375757_15376124 | 0.02 |
Dll1 |
delta like canonical Notch ligand 1 |
36 |
0.97 |
| chr13_41825478_41825636 | 0.02 |
Adtrp |
androgen dependent TFPI regulating protein |
2802 |
0.21 |
| chr5_119332414_119332646 | 0.02 |
n-R5s175 |
nuclear encoded rRNA 5S 175 |
35285 |
0.21 |
| chr5_105743024_105743409 | 0.02 |
Lrrc8d |
leucine rich repeat containing 8D |
10986 |
0.21 |
| chr8_70891500_70891664 | 0.02 |
Slc5a5 |
solute carrier family 5 (sodium iodide symporter), member 5 |
1175 |
0.23 |
| chr5_119669578_119669743 | 0.02 |
Tbx3 |
T-box 3 |
1009 |
0.41 |
| chr6_97311981_97312511 | 0.02 |
Frmd4b |
FERM domain containing 4B |
134 |
0.97 |
| chr1_132800530_132800731 | 0.02 |
Gm22609 |
predicted gene, 22609 |
13718 |
0.19 |
| chr9_122805662_122805885 | 0.02 |
Tcaim |
T cell activation inhibitor, mitochondrial |
170 |
0.92 |
| chr15_25623084_25623317 | 0.02 |
Myo10 |
myosin X |
651 |
0.7 |
| chr8_126474497_126474692 | 0.02 |
Tarbp1 |
TAR RNA binding protein 1 |
471 |
0.78 |
| chr9_36726799_36727144 | 0.02 |
Chek1 |
checkpoint kinase 1 |
94 |
0.95 |
| chr17_47762752_47763230 | 0.02 |
Tfeb |
transcription factor EB |
179 |
0.92 |
| chr1_55087767_55088017 | 0.01 |
Hspd1 |
heat shock protein 1 (chaperonin) |
132 |
0.64 |
| chr10_14174290_14174471 | 0.01 |
Gm28289 |
predicted gene 28289 |
19934 |
0.15 |
| chr3_145758894_145759070 | 0.01 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
307 |
0.91 |
| chr13_44730938_44731098 | 0.01 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
159 |
0.97 |
| chr17_46872155_46872347 | 0.01 |
Bicral |
BRD4 interacting chromatin remodeling complex associated protein like |
13769 |
0.14 |
| chr3_96246696_96246882 | 0.01 |
H3c14 |
H3 clustered histone 14 |
104 |
0.69 |
| chr1_58029738_58029905 | 0.01 |
Aox1 |
aldehyde oxidase 1 |
110 |
0.96 |
| chr8_25754343_25754535 | 0.01 |
Ddhd2 |
DDHD domain containing 2 |
26 |
0.5 |
| chr2_145787645_145787796 | 0.01 |
Rin2 |
Ras and Rab interactor 2 |
1558 |
0.46 |
| chr13_112463928_112464102 | 0.01 |
Il6st |
interleukin 6 signal transducer |
55 |
0.97 |
| chr5_136965885_136966036 | 0.01 |
Cldn15 |
claudin 15 |
656 |
0.49 |
| chr9_48836173_48836358 | 0.01 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
43 |
0.98 |
| chr12_73584355_73584506 | 0.01 |
Prkch |
protein kinase C, eta |
366 |
0.87 |
| chr18_35810644_35810817 | 0.01 |
Ube2d2a |
ubiquitin-conjugating enzyme E2D 2A |
9423 |
0.1 |
| chr4_149998934_149999412 | 0.01 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
2092 |
0.26 |
| chr15_59314887_59315038 | 0.01 |
Sqle |
squalene epoxidase |
115 |
0.96 |
| chr9_44734580_44734731 | 0.01 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
543 |
0.54 |
| chr1_75141569_75141774 | 0.01 |
Cnppd1 |
cyclin Pas1/PHO80 domain containing 1 |
20 |
0.93 |
| chr4_132422119_132422282 | 0.01 |
Phactr4 |
phosphatase and actin regulator 4 |
241 |
0.85 |
| chr7_19676784_19676961 | 0.01 |
Apoc2 |
apolipoprotein C-II |
8 |
0.94 |
| chr2_31125293_31125578 | 0.01 |
Fnbp1 |
formin binding protein 1 |
9123 |
0.16 |
| chr3_90258910_90259076 | 0.01 |
Crtc2 |
CREB regulated transcription coactivator 2 |
28 |
0.94 |
| chr4_149955087_149955244 | 0.01 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
122 |
0.96 |
| chr11_6291801_6292002 | 0.01 |
Ogdh |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
84 |
0.95 |
| chr1_171072220_171072512 | 0.01 |
Gm27552 |
predicted gene, 27552 |
79 |
0.94 |
| chr5_110755870_110756045 | 0.01 |
Gm22583 |
predicted gene, 22583 |
1128 |
0.34 |
| chr4_155216155_155216394 | 0.01 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
6261 |
0.18 |
| chr1_171079830_171080124 | 0.01 |
Gm27844 |
predicted gene, 27844 |
76 |
0.94 |
| chr17_57233819_57233970 | 0.01 |
C3 |
complement component 3 |
5758 |
0.11 |
| chr11_115297531_115297704 | 0.01 |
Fads6 |
fatty acid desaturase domain family, member 6 |
46 |
0.95 |
| chr17_25015240_25015422 | 0.01 |
Cramp1l |
cramped chromatin regulator homolog 1 |
101 |
0.9 |
| chr6_71991498_71991895 | 0.01 |
Gm26628 |
predicted gene, 26628 |
27921 |
0.11 |
| chr12_73908159_73908332 | 0.01 |
Hif1a |
hypoxia inducible factor 1, alpha subunit |
341 |
0.86 |
| chr1_132346178_132346454 | 0.01 |
F730311O21Rik |
RIKEN cDNA F730311O21 gene |
4293 |
0.14 |
| chr17_88064763_88064934 | 0.01 |
Fbxo11 |
F-box protein 11 |
39 |
0.98 |
| chr1_37430023_37430178 | 0.01 |
Coa5 |
cytochrome C oxidase assembly factor 5 |
3 |
0.51 |
| chr19_41483606_41484014 | 0.01 |
Lcor |
ligand dependent nuclear receptor corepressor |
781 |
0.68 |
| chr2_26556400_26556716 | 0.01 |
Gm13358 |
predicted gene 13358 |
2067 |
0.18 |
| chr4_45479517_45479668 | 0.01 |
Shb |
src homology 2 domain-containing transforming protein B |
3210 |
0.21 |
| chr2_167760038_167760213 | 0.01 |
Gm14321 |
predicted gene 14321 |
17342 |
0.13 |
| chr11_68853069_68853241 | 0.01 |
Ndel1 |
nudE neurodevelopment protein 1 like 1 |
20 |
0.97 |
| chr2_29751437_29751588 | 0.01 |
Gm13420 |
predicted gene 13420 |
173 |
0.91 |
| chr3_122483883_122484034 | 0.01 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
235 |
0.9 |
| chr19_26845381_26845541 | 0.01 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
21554 |
0.18 |
| chr8_72322510_72322872 | 0.01 |
Klf2 |
Kruppel-like factor 2 (lung) |
3658 |
0.15 |
| chr11_61855083_61855463 | 0.00 |
Ulk2 |
unc-51 like kinase 2 |
200 |
0.92 |
| chr9_53610323_53610482 | 0.00 |
Acat1 |
acetyl-Coenzyme A acetyltransferase 1 |
20 |
0.97 |
| chr8_70761136_70761327 | 0.00 |
Mpv17l2 |
MPV17 mitochondrial membrane protein-like 2 |
285 |
0.74 |
| chr11_53904286_53904443 | 0.00 |
Gm12218 |
predicted gene 12218 |
1209 |
0.29 |
| chr11_16838911_16839088 | 0.00 |
Egfros |
epidermal growth factor receptor, opposite strand |
8297 |
0.22 |
| chr10_121365310_121365488 | 0.00 |
Gns |
glucosamine (N-acetyl)-6-sulfatase |
223 |
0.9 |
| chr7_81523636_81523823 | 0.00 |
2900076A07Rik |
RIKEN cDNA 2900076A07 gene |
111 |
0.93 |
| chr11_51756413_51756731 | 0.00 |
Sec24a |
Sec24 related gene family, member A (S. cerevisiae) |
46 |
0.97 |
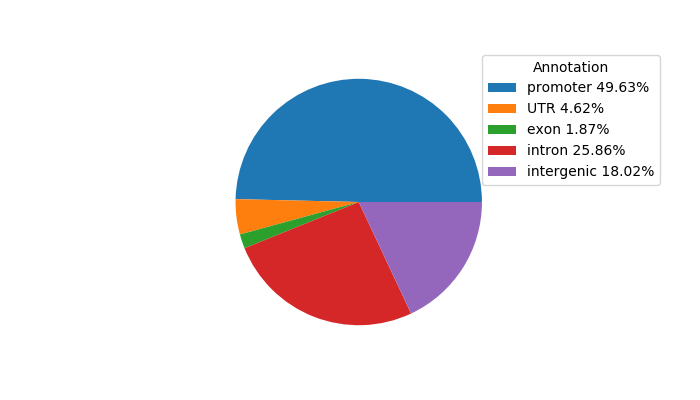
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) histidine transport(GO:0015817) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |