Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
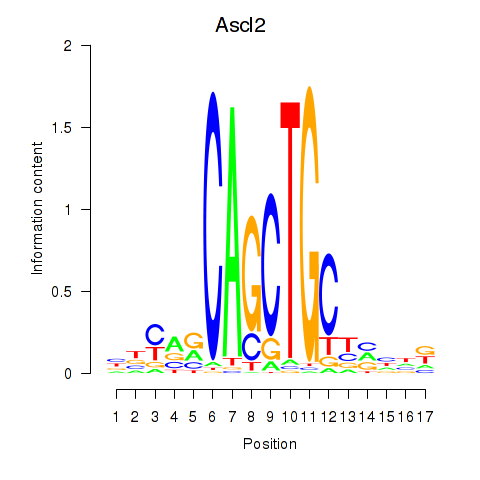
Results for Ascl2
Z-value: 0.53
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSMUSG00000009248.5 | achaete-scute family bHLH transcription factor 2 |
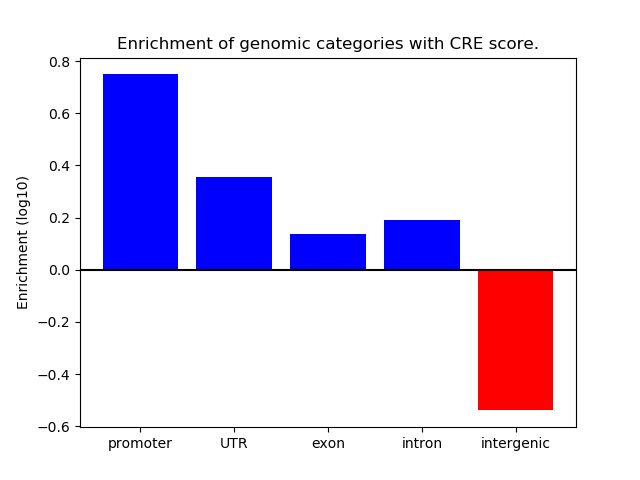
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_142967530_142967799 | Ascl2 | 1053 | 0.436286 | -0.26 | 6.2e-01 | Click! |
Activity of the Ascl2 motif across conditions
Conditions sorted by the z-value of the Ascl2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
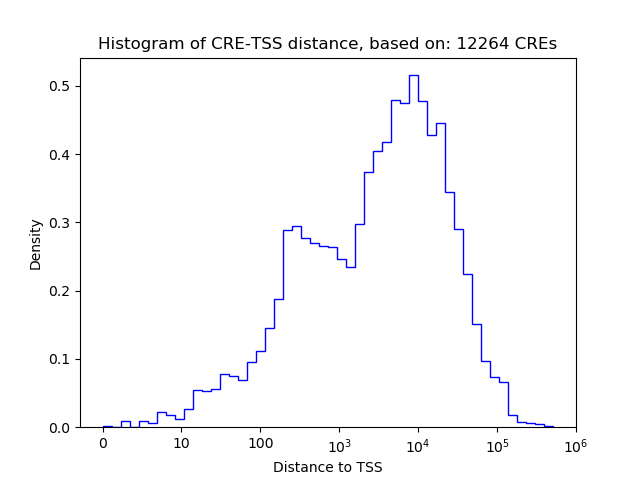
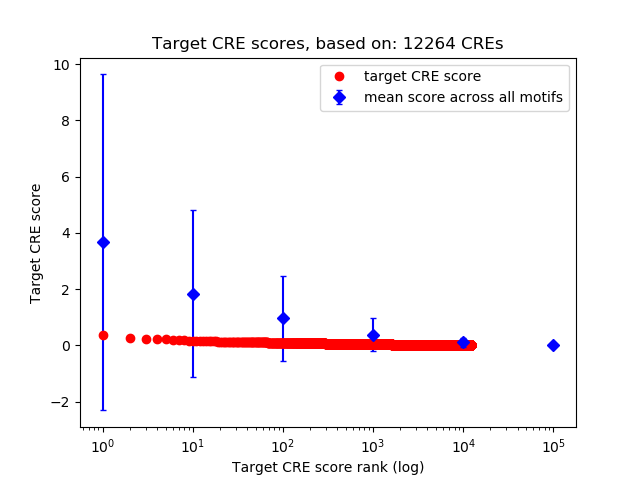
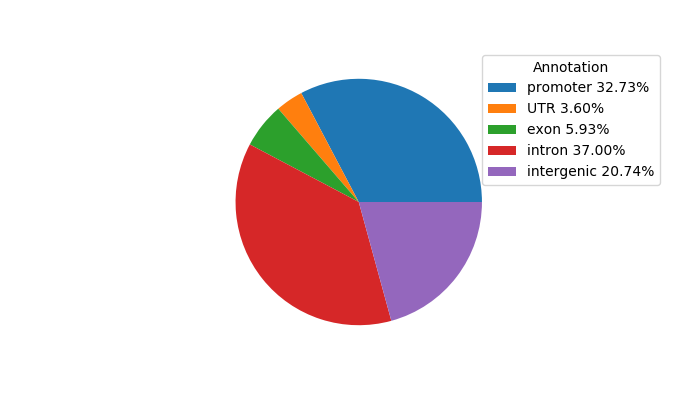
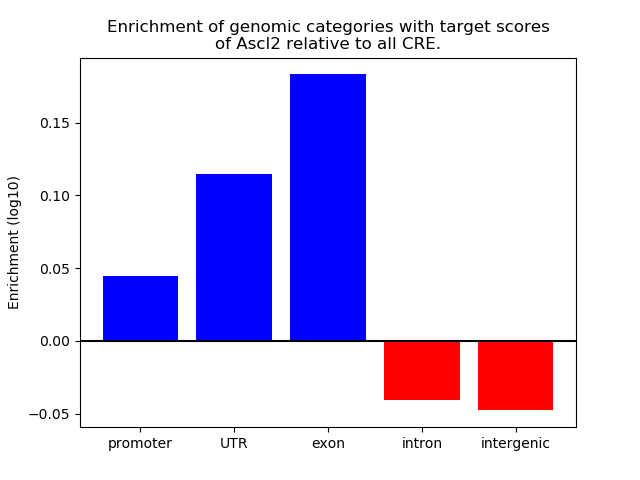
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_67263308_67263719 | 0.38 |
Mlf1 |
myeloid leukemia factor 1 |
110584 |
0.06 |
| chr16_17208938_17209184 | 0.25 |
Rimbp3 |
RIMS binding protein 3 |
458 |
0.65 |
| chr2_173158061_173158212 | 0.23 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
5054 |
0.19 |
| chr10_63410268_63410432 | 0.22 |
Gm7530 |
predicted gene 7530 |
2649 |
0.19 |
| chr14_19751408_19751584 | 0.22 |
Nid2 |
nidogen 2 |
212 |
0.93 |
| chr12_100199636_100199926 | 0.20 |
Calm1 |
calmodulin 1 |
252 |
0.89 |
| chr9_65296934_65297096 | 0.19 |
Gm16218 |
predicted gene 16218 |
904 |
0.39 |
| chr1_160692286_160692437 | 0.18 |
Gm37328 |
predicted gene, 37328 |
34320 |
0.09 |
| chr15_31058451_31058629 | 0.17 |
4930430F21Rik |
RIKEN cDNA 4930430F21 gene |
19412 |
0.2 |
| chr2_173159829_173160580 | 0.16 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
7122 |
0.18 |
| chr2_25262755_25263227 | 0.16 |
Tprn |
taperin |
373 |
0.59 |
| chr11_102399997_102400189 | 0.15 |
Rundc3a |
RUN domain containing 3A |
235 |
0.85 |
| chr1_184873376_184873577 | 0.15 |
C130074G19Rik |
RIKEN cDNA C130074G19 gene |
9742 |
0.16 |
| chr8_94173317_94173477 | 0.15 |
Gm45774 |
predicted gene 45774 |
15 |
0.89 |
| chr8_10937238_10937412 | 0.14 |
Gm45042 |
predicted gene 45042 |
7438 |
0.11 |
| chr10_69208384_69208535 | 0.14 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
93 |
0.97 |
| chr2_31519156_31519420 | 0.14 |
Ass1 |
argininosuccinate synthetase 1 |
798 |
0.61 |
| chr5_114561036_114561196 | 0.14 |
Fam222a |
family with sequence similarity 222, member A |
6900 |
0.17 |
| chr5_124225128_124225319 | 0.14 |
Pitpnm2os1 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 1 |
4502 |
0.13 |
| chr14_65740190_65740357 | 0.13 |
Scara5 |
scavenger receptor class A, member 5 |
63796 |
0.1 |
| chr12_28910370_28910535 | 0.13 |
Gm31508 |
predicted gene, 31508 |
223 |
0.94 |
| chr1_93137985_93138369 | 0.13 |
Agxt |
alanine-glyoxylate aminotransferase |
1702 |
0.26 |
| chr11_120013435_120013644 | 0.13 |
Aatk |
apoptosis-associated tyrosine kinase |
1131 |
0.23 |
| chr5_24976790_24976954 | 0.13 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
8970 |
0.18 |
| chr19_46141233_46141384 | 0.13 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
325 |
0.82 |
| chr1_59157472_59157626 | 0.13 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
1112 |
0.34 |
| chr7_114325255_114325465 | 0.12 |
Psma1 |
proteasome subunit alpha 1 |
49242 |
0.13 |
| chr3_116329378_116329892 | 0.12 |
Gm29151 |
predicted gene 29151 |
20468 |
0.17 |
| chr1_36149451_36149602 | 0.12 |
Uggt1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
998 |
0.45 |
| chr12_84187018_84187182 | 0.12 |
Gm19327 |
predicted gene, 19327 |
706 |
0.52 |
| chr12_28800275_28800436 | 0.12 |
Gm48905 |
predicted gene, 48905 |
9783 |
0.16 |
| chr19_3893794_3894051 | 0.12 |
Chka |
choline kinase alpha |
2191 |
0.13 |
| chr9_47324524_47324675 | 0.12 |
Gm31816 |
predicted gene, 31816 |
38151 |
0.19 |
| chr4_117040674_117041081 | 0.12 |
Eif2b3 |
eukaryotic translation initiation factor 2B, subunit 3 |
3599 |
0.12 |
| chr3_108788010_108788161 | 0.12 |
Stxbp3 |
syntaxin binding protein 3 |
9699 |
0.15 |
| chr19_32648548_32648744 | 0.12 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
28641 |
0.18 |
| chr2_25472167_25472318 | 0.12 |
Ptgds |
prostaglandin D2 synthase (brain) |
2196 |
0.12 |
| chr4_82415983_82416134 | 0.11 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
23352 |
0.23 |
| chr19_45749067_45749428 | 0.11 |
Gm15491 |
predicted gene 15491 |
261 |
0.52 |
| chr10_75783826_75784124 | 0.11 |
Gstt3 |
glutathione S-transferase, theta 3 |
2561 |
0.13 |
| chr13_74209069_74209298 | 0.11 |
Exoc3 |
exocyst complex component 3 |
451 |
0.81 |
| chr19_46628011_46628401 | 0.11 |
Wbp1l |
WW domain binding protein 1 like |
4805 |
0.15 |
| chrX_63961215_63961366 | 0.11 |
Gm8260 |
predicted gene 8260 |
86616 |
0.1 |
| chr7_30051105_30051256 | 0.11 |
Zfp14 |
zinc finger protein 14 |
193 |
0.88 |
| chr13_93628549_93628834 | 0.11 |
Gm15622 |
predicted gene 15622 |
3309 |
0.2 |
| chr17_31306868_31307019 | 0.11 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
522 |
0.71 |
| chr17_56108904_56109250 | 0.11 |
Plin4 |
perilipin 4 |
725 |
0.45 |
| chr2_27212850_27213071 | 0.11 |
Sardh |
sarcosine dehydrogenase |
2672 |
0.2 |
| chrX_169904110_169904261 | 0.11 |
Mid1 |
midline 1 |
22766 |
0.2 |
| chr11_35458160_35458335 | 0.11 |
Slit3 |
slit guidance ligand 3 |
86438 |
0.09 |
| chr6_149138694_149138910 | 0.11 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
1047 |
0.41 |
| chr14_12103232_12103447 | 0.11 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
12179 |
0.23 |
| chr17_49440803_49440954 | 0.11 |
Mocs1 |
molybdenum cofactor synthesis 1 |
7682 |
0.22 |
| chr12_112597485_112597658 | 0.11 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
7086 |
0.14 |
| chr4_141663419_141663714 | 0.11 |
Plekhm2 |
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
552 |
0.67 |
| chr6_72120521_72121047 | 0.11 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr10_24915615_24916009 | 0.10 |
Arg1 |
arginase, liver |
595 |
0.65 |
| chr7_112276934_112277125 | 0.10 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
5730 |
0.31 |
| chr5_122060223_122060392 | 0.10 |
Cux2 |
cut-like homeobox 2 |
10205 |
0.14 |
| chr8_104909005_104909158 | 0.10 |
Gm8798 |
predicted gene 8798 |
16335 |
0.09 |
| chr10_80444503_80444654 | 0.10 |
Tcf3 |
transcription factor 3 |
10931 |
0.09 |
| chr9_107738378_107738568 | 0.10 |
Rbm5 |
RNA binding motif protein 5 |
5761 |
0.11 |
| chr9_52056529_52056697 | 0.10 |
Rdx |
radixin |
8342 |
0.19 |
| chr8_104443177_104443346 | 0.10 |
Dync1li2 |
dynein, cytoplasmic 1 light intermediate chain 2 |
214 |
0.83 |
| chr2_31513768_31514291 | 0.10 |
Ass1 |
argininosuccinate synthetase 1 |
4461 |
0.2 |
| chr6_145606729_145606899 | 0.10 |
Lmntd1 |
lamin tail domain containing 1 |
7155 |
0.21 |
| chr17_25068673_25068863 | 0.10 |
Tmem204 |
transmembrane protein 204 |
9456 |
0.11 |
| chr2_180788221_180788717 | 0.10 |
Bhlhe23 |
basic helix-loop-helix family, member e23 |
11569 |
0.11 |
| chrX_108664177_108664328 | 0.10 |
Gm379 |
predicted gene 379 |
203 |
0.97 |
| chr8_68277116_68277418 | 0.10 |
Sh2d4a |
SH2 domain containing 4A |
700 |
0.69 |
| chr17_34733622_34734232 | 0.10 |
C4b |
complement component 4B (Chido blood group) |
2172 |
0.12 |
| chr2_31491761_31492352 | 0.10 |
Ass1 |
argininosuccinate synthetase 1 |
5716 |
0.2 |
| chr2_167148083_167148259 | 0.10 |
1110018N20Rik |
RIKEN cDNA 1110018N20 gene |
40531 |
0.08 |
| chr8_93270889_93271065 | 0.10 |
Ces1f |
carboxylesterase 1F |
3572 |
0.18 |
| chr5_121761085_121761315 | 0.10 |
Atxn2 |
ataxin 2 |
12004 |
0.12 |
| chr7_30943687_30943953 | 0.10 |
Hamp |
hepcidin antimicrobial peptide |
212 |
0.82 |
| chr11_69122450_69122661 | 0.10 |
9130213A22Rik |
RIKEN cDNA 9130213A22 gene |
34 |
0.93 |
| chr8_124273757_124274124 | 0.10 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
21163 |
0.15 |
| chr14_120534643_120535027 | 0.10 |
Rap2a |
RAS related protein 2a |
56391 |
0.14 |
| chr5_8988023_8988336 | 0.10 |
Crot |
carnitine O-octanoyltransferase |
6035 |
0.13 |
| chr15_75996647_75996807 | 0.10 |
Mapk15 |
mitogen-activated protein kinase 15 |
14 |
0.94 |
| chr14_8251423_8251762 | 0.10 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
1930 |
0.34 |
| chr2_167877367_167877523 | 0.10 |
Gm14319 |
predicted gene 14319 |
18860 |
0.16 |
| chr16_11034212_11034466 | 0.10 |
Gm10832 |
predicted gene 10832 |
2383 |
0.16 |
| chr17_28279922_28280073 | 0.09 |
Ppard |
peroxisome proliferator activator receptor delta |
7878 |
0.11 |
| chr7_28732426_28732636 | 0.09 |
Fbxo17 |
F-box protein 17 |
31 |
0.95 |
| chr6_117178128_117178348 | 0.09 |
Cxcl12 |
chemokine (C-X-C motif) ligand 12 |
9661 |
0.18 |
| chr9_77964186_77964368 | 0.09 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
22986 |
0.11 |
| chr6_29539642_29539820 | 0.09 |
Irf5 |
interferon regulatory factor 5 |
1415 |
0.29 |
| chr1_190164468_190164683 | 0.09 |
Gm28172 |
predicted gene 28172 |
4095 |
0.21 |
| chr13_96661137_96661370 | 0.09 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
4586 |
0.17 |
| chr11_109616366_109616667 | 0.09 |
Gm11685 |
predicted gene 11685 |
196 |
0.93 |
| chr8_126498607_126498804 | 0.09 |
Gm6091 |
predicted pseudogene 6091 |
22293 |
0.18 |
| chr2_180756949_180757112 | 0.09 |
Gm14343 |
predicted gene 14343 |
18373 |
0.1 |
| chr2_48411517_48411668 | 0.09 |
Gm13472 |
predicted gene 13472 |
23555 |
0.19 |
| chr16_23974024_23974210 | 0.09 |
Bcl6 |
B cell leukemia/lymphoma 6 |
1737 |
0.33 |
| chr13_37761078_37761249 | 0.09 |
Gm31600 |
predicted gene, 31600 |
585 |
0.68 |
| chr12_108292499_108292690 | 0.09 |
Hhipl1 |
hedgehog interacting protein-like 1 |
13676 |
0.15 |
| chr16_67621078_67621243 | 0.09 |
Cadm2 |
cell adhesion molecule 2 |
252 |
0.95 |
| chr11_119969786_119969965 | 0.09 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
9854 |
0.11 |
| chr11_7136281_7136476 | 0.09 |
Adcy1 |
adenylate cyclase 1 |
23504 |
0.18 |
| chr2_173161537_173161720 | 0.09 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
8546 |
0.17 |
| chr11_120012134_120012364 | 0.09 |
Aatk |
apoptosis-associated tyrosine kinase |
159 |
0.9 |
| chr5_8962841_8962998 | 0.09 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
4367 |
0.15 |
| chr6_22037034_22037204 | 0.09 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
11176 |
0.27 |
| chr11_97449921_97450124 | 0.09 |
Arhgap23 |
Rho GTPase activating protein 23 |
138 |
0.95 |
| chr1_39070328_39070509 | 0.09 |
Gm37821 |
predicted gene, 37821 |
12781 |
0.17 |
| chr4_44994891_44995176 | 0.09 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
13533 |
0.1 |
| chr9_67842866_67843237 | 0.09 |
Vps13c |
vacuolar protein sorting 13C |
2639 |
0.27 |
| chr18_46651026_46651177 | 0.09 |
Gm3734 |
predicted gene 3734 |
20294 |
0.14 |
| chr19_56652542_56652727 | 0.09 |
Gm32441 |
predicted gene, 32441 |
11977 |
0.17 |
| chr1_67195779_67195930 | 0.09 |
Gm15668 |
predicted gene 15668 |
53346 |
0.13 |
| chr3_152478706_152478877 | 0.09 |
Ak5 |
adenylate kinase 5 |
276 |
0.91 |
| chr19_3451534_3452133 | 0.09 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
19172 |
0.12 |
| chr4_137795816_137796289 | 0.09 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
178 |
0.96 |
| chr7_141335382_141335533 | 0.09 |
Tmem80 |
transmembrane protein 80 |
1799 |
0.16 |
| chr4_63494908_63495098 | 0.09 |
Whrn |
whirlin |
244 |
0.91 |
| chr19_37509643_37509826 | 0.09 |
Exoc6 |
exocyst complex component 6 |
11145 |
0.17 |
| chr9_83834067_83834712 | 0.09 |
Ttk |
Ttk protein kinase |
300 |
0.91 |
| chr8_90834012_90834186 | 0.09 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
5243 |
0.14 |
| chr9_110985950_110986443 | 0.09 |
1700061E17Rik |
RIKEN cDNA 1700061E17 gene |
5 |
0.94 |
| chr6_72360494_72360859 | 0.09 |
Rnf181 |
ring finger protein 181 |
391 |
0.72 |
| chr2_26593869_26594069 | 0.09 |
Egfl7 |
EGF-like domain 7 |
1822 |
0.16 |
| chr2_103472788_103473367 | 0.09 |
Cat |
catalase |
12048 |
0.18 |
| chr15_79002341_79002492 | 0.09 |
Mir6956 |
microRNA 6956 |
193 |
0.52 |
| chr19_42602482_42602861 | 0.09 |
Loxl4 |
lysyl oxidase-like 4 |
650 |
0.71 |
| chr11_117964790_117965236 | 0.09 |
Socs3 |
suppressor of cytokine signaling 3 |
4173 |
0.16 |
| chr10_122454632_122454783 | 0.09 |
Gm48878 |
predicted gene, 48878 |
575 |
0.76 |
| chr5_140389137_140389453 | 0.09 |
Snx8 |
sorting nexin 8 |
33 |
0.97 |
| chr2_32414031_32414217 | 0.09 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
2447 |
0.14 |
| chr8_106606041_106606388 | 0.09 |
Cdh1 |
cadherin 1 |
2071 |
0.29 |
| chr5_120483296_120483855 | 0.09 |
Gm15690 |
predicted gene 15690 |
2672 |
0.15 |
| chr3_75950402_75950574 | 0.09 |
Gm37685 |
predicted gene, 37685 |
1636 |
0.35 |
| chr12_112675159_112675325 | 0.09 |
Akt1 |
thymoma viral proto-oncogene 1 |
358 |
0.75 |
| chr3_86002499_86002650 | 0.09 |
Prss48 |
protease, serine 48 |
83 |
0.96 |
| chr8_123653291_123653442 | 0.09 |
Rhou |
ras homolog family member U |
563 |
0.38 |
| chr2_93477672_93477838 | 0.09 |
Gm10804 |
predicted gene 10804 |
9316 |
0.19 |
| chr5_87080846_87081015 | 0.09 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
10227 |
0.11 |
| chr2_122775480_122775660 | 0.08 |
Sqor |
sulfide quinone oxidoreductase |
3852 |
0.22 |
| chr11_102214881_102215057 | 0.08 |
Hdac5 |
histone deacetylase 5 |
3959 |
0.11 |
| chr8_46512530_46512928 | 0.08 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
14057 |
0.15 |
| chr14_34675574_34675754 | 0.08 |
Wapl |
WAPL cohesin release factor |
1491 |
0.26 |
| chr6_72233008_72233332 | 0.08 |
Atoh8 |
atonal bHLH transcription factor 8 |
1367 |
0.39 |
| chr3_69403326_69403495 | 0.08 |
Gm17213 |
predicted gene 17213 |
28958 |
0.17 |
| chr11_85846806_85847022 | 0.08 |
Gm11444 |
predicted gene 11444 |
3419 |
0.17 |
| chr3_84639824_84639982 | 0.08 |
Tmem154 |
transmembrane protein 154 |
26289 |
0.17 |
| chr11_78157696_78157847 | 0.08 |
Traf4 |
TNF receptor associated factor 4 |
7745 |
0.07 |
| chr13_96749686_96749877 | 0.08 |
Ankrd31 |
ankyrin repeat domain 31 |
1249 |
0.45 |
| chr17_56207962_56208313 | 0.08 |
Dpp9 |
dipeptidylpeptidase 9 |
1071 |
0.32 |
| chr3_67262698_67262957 | 0.08 |
Mlf1 |
myeloid leukemia factor 1 |
111270 |
0.06 |
| chr5_33901574_33901762 | 0.08 |
Mir7024 |
microRNA 7024 |
2697 |
0.18 |
| chr12_84221379_84221549 | 0.08 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
2583 |
0.16 |
| chr7_121907300_121907714 | 0.08 |
Scnn1b |
sodium channel, nonvoltage-gated 1 beta |
8279 |
0.18 |
| chr16_18069271_18069422 | 0.08 |
Dgcr6 |
DiGeorge syndrome critical region gene 6 |
134 |
0.94 |
| chr13_7414710_7414870 | 0.08 |
Gm36074 |
predicted gene, 36074 |
31719 |
0.23 |
| chr5_93250297_93250457 | 0.08 |
Ccng2 |
cyclin G2 |
16880 |
0.14 |
| chr5_115575131_115575282 | 0.08 |
Mir7029 |
microRNA 7029 |
5743 |
0.11 |
| chr2_163551700_163552131 | 0.08 |
Hnf4a |
hepatic nuclear factor 4, alpha |
1832 |
0.25 |
| chr5_123977607_123977811 | 0.08 |
Hip1r |
huntingtin interacting protein 1 related |
2545 |
0.16 |
| chr8_14257136_14257302 | 0.08 |
Gm26184 |
predicted gene, 26184 |
16092 |
0.22 |
| chr1_134955632_134955799 | 0.08 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
136 |
0.96 |
| chr1_36512638_36512794 | 0.08 |
Cnnm3 |
cyclin M3 |
381 |
0.72 |
| chr2_32678870_32679034 | 0.08 |
Gm26236 |
predicted gene, 26236 |
3843 |
0.08 |
| chr5_114606575_114606726 | 0.08 |
Trpv4 |
transient receptor potential cation channel, subfamily V, member 4 |
23988 |
0.13 |
| chr2_110007906_110008057 | 0.08 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
7453 |
0.19 |
| chr18_46718177_46718358 | 0.08 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
9762 |
0.13 |
| chr15_4727530_4727980 | 0.08 |
C6 |
complement component 6 |
539 |
0.84 |
| chr8_40187640_40187799 | 0.08 |
Fgf20 |
fibroblast growth factor 20 |
99234 |
0.07 |
| chr8_121573324_121573508 | 0.08 |
Fbxo31 |
F-box protein 31 |
5331 |
0.12 |
| chr19_45036974_45037129 | 0.08 |
Pdzd7 |
PDZ domain containing 7 |
7036 |
0.1 |
| chr11_101070404_101070591 | 0.08 |
Naglu |
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
485 |
0.62 |
| chr18_4970911_4971104 | 0.08 |
Svil |
supervillin |
22806 |
0.26 |
| chr1_74047431_74047619 | 0.08 |
Tns1 |
tensin 1 |
10368 |
0.21 |
| chr3_129970893_129971067 | 0.08 |
Mcub |
mitochondrial calcium uniporter dominant negative beta subunit |
774 |
0.62 |
| chr11_115608626_115608777 | 0.08 |
Mif4gd |
MIF4G domain containing |
3425 |
0.11 |
| chr17_12418655_12418810 | 0.08 |
Plg |
plasminogen |
40073 |
0.12 |
| chr15_102006414_102006639 | 0.08 |
Krt8 |
keratin 8 |
2044 |
0.19 |
| chr6_133105397_133105578 | 0.08 |
Smim10l1 |
small integral membrane protein 10 like 1 |
5 |
0.95 |
| chr10_19525625_19525776 | 0.08 |
Gm48563 |
predicted gene, 48563 |
3050 |
0.27 |
| chr8_70182055_70182225 | 0.08 |
Tmem161a |
transmembrane protein 161A |
1601 |
0.22 |
| chr3_84647383_84647687 | 0.08 |
Tmem154 |
transmembrane protein 154 |
18657 |
0.2 |
| chr2_32727754_32727916 | 0.08 |
Sh2d3c |
SH2 domain containing 3C |
129 |
0.89 |
| chr15_67116763_67117052 | 0.08 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
2915 |
0.34 |
| chr14_30922623_30922783 | 0.08 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
846 |
0.45 |
| chr10_94055869_94056059 | 0.08 |
Fgd6 |
FYVE, RhoGEF and PH domain containing 6 |
16578 |
0.11 |
| chr1_182609269_182609652 | 0.08 |
Capn8 |
calpain 8 |
44422 |
0.12 |
| chr7_78782525_78782681 | 0.08 |
Mrpl46 |
mitochondrial ribosomal protein L46 |
486 |
0.44 |
| chr8_111745534_111745719 | 0.08 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
1817 |
0.34 |
| chr6_72117206_72117587 | 0.08 |
St3gal5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
2164 |
0.2 |
| chr2_158768137_158768288 | 0.07 |
Fam83d |
family with sequence similarity 83, member D |
119 |
0.96 |
| chr15_75981322_75981497 | 0.07 |
Ccdc166 |
coiled-coil domain containing 166 |
1033 |
0.28 |
| chr13_37778325_37778511 | 0.07 |
Rreb1 |
ras responsive element binding protein 1 |
18 |
0.97 |
| chr18_75818786_75819091 | 0.07 |
Zbtb7c |
zinc finger and BTB domain containing 7C |
1240 |
0.52 |
| chr9_113127551_113127725 | 0.07 |
Gm36251 |
predicted gene, 36251 |
4609 |
0.31 |
| chr12_8000122_8000489 | 0.07 |
Apob |
apolipoprotein B |
12054 |
0.22 |
| chr17_31908788_31908992 | 0.07 |
2310015A16Rik |
RIKEN cDNA 2310015A16 gene |
139 |
0.94 |
| chr10_94514734_94514917 | 0.07 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
32 |
0.98 |
| chr1_164210089_164210240 | 0.07 |
Slc19a2 |
solute carrier family 19 (thiamine transporter), member 2 |
38882 |
0.1 |
| chr4_48125260_48125479 | 0.07 |
Stx17 |
syntaxin 17 |
434 |
0.87 |
| chrX_74023375_74023542 | 0.07 |
Irak1 |
interleukin-1 receptor-associated kinase 1 |
50 |
0.58 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.0 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.0 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.0 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |