Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
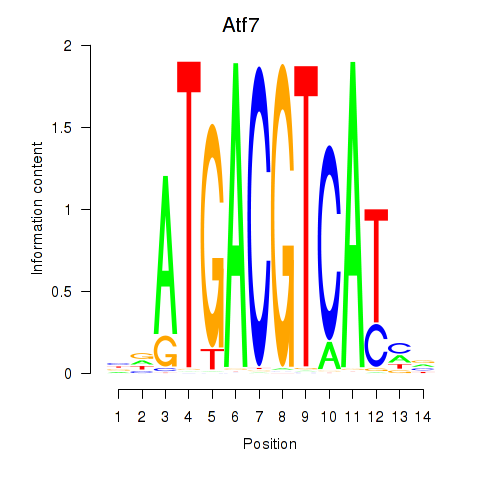
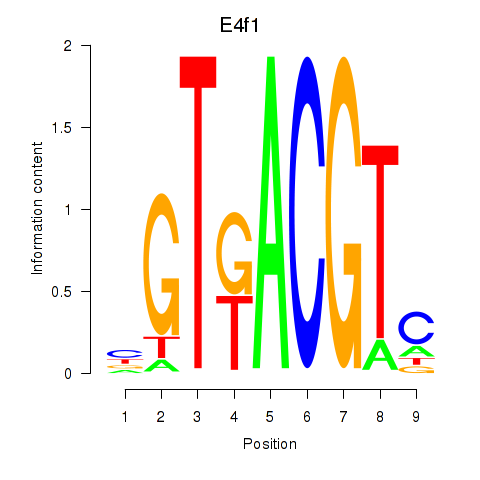
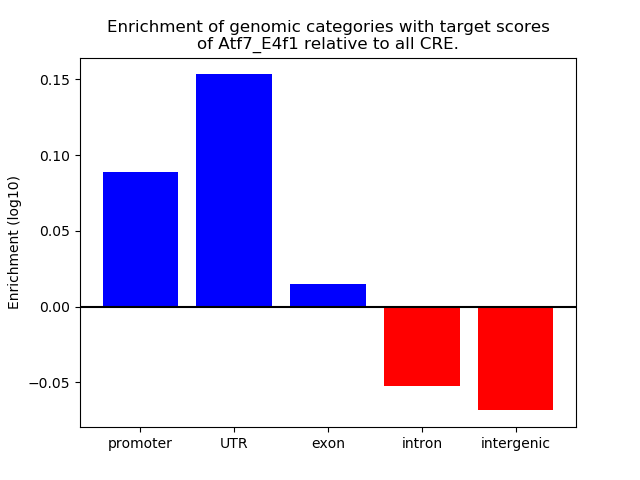
Results for Atf7_E4f1
Z-value: 0.77
Transcription factors associated with Atf7_E4f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf7
|
ENSMUSG00000052414.9 | activating transcription factor 7 |
|
Atf7
|
ENSMUSG00000071584.1 | activating transcription factor 7 |
|
E4f1
|
ENSMUSG00000024137.8 | E4F transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_24469959_24470159 | E4f1 | 132 | 0.699941 | -0.92 | 9.9e-03 | Click! |
| chr17_24470160_24470458 | E4f1 | 4 | 0.544263 | -0.84 | 3.7e-02 | Click! |
| chr17_24453368_24453643 | E4f1 | 1781 | 0.146839 | 0.75 | 8.4e-02 | Click! |
| chr17_24455257_24455550 | E4f1 | 11 | 0.934138 | -0.67 | 1.4e-01 | Click! |
| chr17_24454731_24454905 | E4f1 | 468 | 0.582543 | -0.67 | 1.5e-01 | Click! |
Activity of the Atf7_E4f1 motif across conditions
Conditions sorted by the z-value of the Atf7_E4f1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
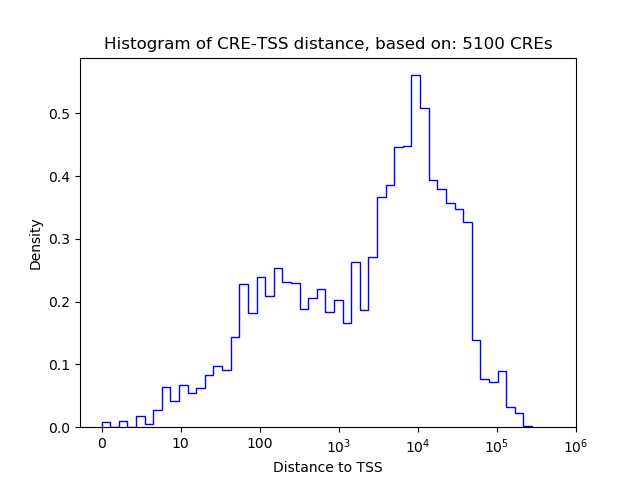
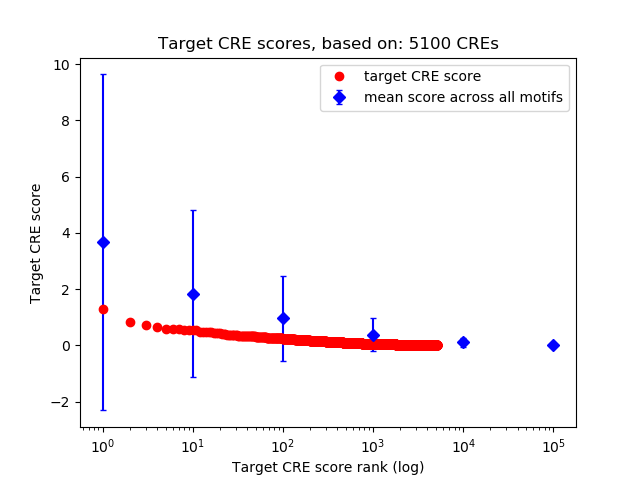
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_75173089_75173918 | 1.28 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr3_51255928_51256079 | 0.83 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr4_141858646_141858827 | 0.71 |
Ctrc |
chymotrypsin C (caldecrin) |
12377 |
0.1 |
| chr3_146630472_146630703 | 0.64 |
Gm16325 |
predicted gene 16325 |
11345 |
0.12 |
| chr8_40881703_40881854 | 0.57 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
6830 |
0.17 |
| chr7_97788226_97788577 | 0.57 |
Pak1 |
p21 (RAC1) activated kinase 1 |
140 |
0.96 |
| chr2_156065132_156065865 | 0.56 |
Spag4 |
sperm associated antigen 4 |
55 |
0.94 |
| chr9_42464439_42464590 | 0.55 |
Tbcel |
tubulin folding cofactor E-like |
3053 |
0.23 |
| chr10_17948008_17948419 | 0.55 |
Heca |
hdc homolog, cell cycle regulator |
146 |
0.97 |
| chr9_55264900_55265092 | 0.54 |
Nrg4 |
neuregulin 4 |
18576 |
0.16 |
| chr3_27751423_27751574 | 0.53 |
Fndc3b |
fibronectin type III domain containing 3B |
40191 |
0.19 |
| chr19_61226929_61227456 | 0.49 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr18_74939611_74939786 | 0.49 |
Gm18786 |
predicted gene, 18786 |
3418 |
0.13 |
| chr14_61680870_61681697 | 0.47 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr2_180589607_180589819 | 0.47 |
Ogfr |
opioid growth factor receptor |
251 |
0.89 |
| chr12_32766682_32766844 | 0.46 |
Nampt |
nicotinamide phosphoribosyltransferase |
52782 |
0.12 |
| chr15_102103356_102103597 | 0.44 |
Tns2 |
tensin 2 |
488 |
0.69 |
| chr17_70668731_70668893 | 0.43 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
86770 |
0.08 |
| chr2_72741614_72741840 | 0.43 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
9126 |
0.22 |
| chr4_101741736_101741946 | 0.42 |
Lepr |
leptin receptor |
24434 |
0.19 |
| chr5_92607049_92607221 | 0.39 |
Stbd1 |
starch binding domain 1 |
4067 |
0.19 |
| chr11_16852969_16853170 | 0.39 |
Egfros |
epidermal growth factor receptor, opposite strand |
22367 |
0.17 |
| chr10_69204774_69204937 | 0.38 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
3697 |
0.22 |
| chr7_97757551_97757739 | 0.38 |
Aqp11 |
aquaporin 11 |
19356 |
0.15 |
| chr15_59032105_59032256 | 0.37 |
Mtss1 |
MTSS I-BAR domain containing 1 |
8416 |
0.21 |
| chr2_8124301_8124468 | 0.37 |
Gm13254 |
predicted gene 13254 |
23481 |
0.29 |
| chr2_155070811_155070993 | 0.36 |
Gm45609 |
predicted gene 45609 |
3279 |
0.17 |
| chr14_120477462_120478025 | 0.36 |
Rap2a |
RAS related protein 2a |
701 |
0.77 |
| chr14_102015145_102015320 | 0.36 |
Gm5854 |
predicted gene 5854 |
32469 |
0.22 |
| chr1_54492445_54492606 | 0.35 |
Gm36955 |
predicted gene, 36955 |
27745 |
0.14 |
| chr15_58972603_58972890 | 0.35 |
Mtss1 |
MTSS I-BAR domain containing 1 |
207 |
0.92 |
| chr18_62095425_62095728 | 0.35 |
Gm41750 |
predicted gene, 41750 |
47042 |
0.14 |
| chr3_102677050_102677222 | 0.34 |
Gm19202 |
predicted gene, 19202 |
9581 |
0.14 |
| chr6_90599935_90600097 | 0.34 |
Gm15756 |
predicted gene 15756 |
68 |
0.96 |
| chr16_96127404_96127899 | 0.34 |
Hmgn1 |
high mobility group nucleosomal binding domain 1 |
47 |
0.96 |
| chr7_118704359_118704549 | 0.34 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
926 |
0.45 |
| chr10_89492629_89492900 | 0.34 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
13885 |
0.21 |
| chr10_86885775_86886048 | 0.34 |
Stab2 |
stabilin 2 |
162 |
0.93 |
| chr7_19933617_19933791 | 0.34 |
Igsf23 |
immunoglobulin superfamily, member 23 |
11194 |
0.07 |
| chr9_106238027_106238241 | 0.33 |
Alas1 |
aminolevulinic acid synthase 1 |
596 |
0.58 |
| chr11_16850729_16850984 | 0.33 |
Egfros |
epidermal growth factor receptor, opposite strand |
20154 |
0.18 |
| chrY_90783982_90784262 | 0.33 |
Gm47283 |
predicted gene, 47283 |
616 |
0.72 |
| chr9_48596792_48597151 | 0.33 |
Nnmt |
nicotinamide N-methyltransferase |
404 |
0.88 |
| chr11_3443162_3443363 | 0.33 |
Rnf185 |
ring finger protein 185 |
9072 |
0.1 |
| chr3_138288275_138288480 | 0.33 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
10726 |
0.12 |
| chr17_27820677_27821072 | 0.32 |
Ilrun |
inflammation and lipid regulator with UBA-like and NBR1-like domains |
226 |
0.89 |
| chr6_27782346_27782517 | 0.32 |
Gm26310 |
predicted gene, 26310 |
114344 |
0.07 |
| chr10_87926267_87926684 | 0.32 |
Tyms-ps |
thymidylate synthase, pseudogene |
40372 |
0.13 |
| chr2_9990244_9990418 | 0.31 |
Taf3 |
TATA-box binding protein associated factor 3 |
36674 |
0.1 |
| chr10_80622017_80622180 | 0.31 |
Csnk1g2 |
casein kinase 1, gamma 2 |
740 |
0.4 |
| chr5_138863743_138864096 | 0.31 |
Gm5294 |
predicted gene 5294 |
43839 |
0.13 |
| chr4_60380015_60380268 | 0.31 |
Mup-ps29 |
major urinary protein, pseudogene 29 |
4353 |
0.18 |
| chr13_109536856_109537007 | 0.31 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
44474 |
0.21 |
| chr2_52603393_52603554 | 0.30 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
16281 |
0.2 |
| chr15_85599225_85599378 | 0.30 |
Wnt7b |
wingless-type MMTV integration site family, member 7B |
16828 |
0.15 |
| chr4_61740192_61740453 | 0.29 |
Gm12573 |
predicted gene 12573 |
9753 |
0.14 |
| chr15_12321006_12321530 | 0.29 |
Golph3 |
golgi phosphoprotein 3 |
182 |
0.84 |
| chr5_135685581_135685753 | 0.29 |
Por |
P450 (cytochrome) oxidoreductase |
3369 |
0.13 |
| chr4_61401423_61401691 | 0.29 |
Mup15 |
major urinary protein 15 |
38186 |
0.14 |
| chr14_69411185_69411411 | 0.29 |
Gm38378 |
predicted gene, 38378 |
13527 |
0.11 |
| chr4_149854310_149854480 | 0.28 |
Gm47301 |
predicted gene, 47301 |
36631 |
0.09 |
| chr3_148681139_148681436 | 0.28 |
Gm43575 |
predicted gene 43575 |
28227 |
0.24 |
| chr8_111145184_111145817 | 0.28 |
9430091E24Rik |
RIKEN cDNA 9430091E24 gene |
20 |
0.97 |
| chr8_111630041_111630248 | 0.28 |
Ldhd |
lactate dehydrogenase D |
191 |
0.93 |
| chr10_75783826_75784124 | 0.28 |
Gstt3 |
glutathione S-transferase, theta 3 |
2561 |
0.13 |
| chr4_139198021_139198318 | 0.27 |
Capzb |
capping protein (actin filament) muscle Z-line, beta |
1098 |
0.44 |
| chr7_139388680_139389161 | 0.27 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
189 |
0.96 |
| chr15_80836341_80836511 | 0.27 |
Tnrc6b |
trinucleotide repeat containing 6b |
37711 |
0.15 |
| chr14_75242017_75242184 | 0.27 |
Cpb2 |
carboxypeptidase B2 (plasma) |
187 |
0.93 |
| chr3_10439906_10440215 | 0.27 |
Snx16 |
sorting nexin 16 |
27 |
0.98 |
| chr9_21367416_21367785 | 0.26 |
Ilf3 |
interleukin enhancer binding factor 3 |
271 |
0.81 |
| chr15_58941890_58942087 | 0.26 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
8180 |
0.13 |
| chr2_102154219_102154407 | 0.26 |
Ldlrad3 |
low density lipoprotein receptor class A domain containing 3 |
16194 |
0.19 |
| chr2_58780980_58781131 | 0.26 |
Upp2 |
uridine phosphorylase 2 |
15730 |
0.2 |
| chr16_31521897_31522062 | 0.26 |
Gm46560 |
predicted gene, 46560 |
13592 |
0.16 |
| chr12_59219708_59220018 | 0.26 |
Fbxo33 |
F-box protein 33 |
138 |
0.95 |
| chr11_49837892_49838056 | 0.26 |
Mapk9 |
mitogen-activated protein kinase 9 |
8777 |
0.16 |
| chr11_88242033_88242214 | 0.26 |
Gm38534 |
predicted gene, 38534 |
11245 |
0.17 |
| chr2_79448009_79448160 | 0.26 |
Neurod1 |
neurogenic differentiation 1 |
8667 |
0.21 |
| chr3_51234740_51234908 | 0.25 |
Gm38357 |
predicted gene, 38357 |
2907 |
0.19 |
| chr6_145855716_145856207 | 0.25 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr4_11966482_11966677 | 0.25 |
1700123M08Rik |
RIKEN cDNA 1700123M08 gene |
5 |
0.62 |
| chr14_67072405_67072747 | 0.25 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
132 |
0.95 |
| chr4_102432201_102432362 | 0.25 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
2234 |
0.46 |
| chr2_57114946_57115097 | 0.25 |
Nr4a2 |
nuclear receptor subfamily 4, group A, member 2 |
17 |
0.98 |
| chr4_84540692_84540849 | 0.25 |
Bnc2 |
basonuclin 2 |
5520 |
0.32 |
| chr8_70302451_70302610 | 0.25 |
Cope |
coatomer protein complex, subunit epsilon |
12 |
0.5 |
| chr10_124731285_124731709 | 0.25 |
Gm47839 |
predicted gene, 47839 |
113984 |
0.07 |
| chr9_23224794_23225147 | 0.25 |
Bmper |
BMP-binding endothelial regulator |
1684 |
0.55 |
| chr10_20952125_20952679 | 0.25 |
Ahi1 |
Abelson helper integration site 1 |
145 |
0.97 |
| chr4_141060479_141060918 | 0.25 |
Crocc |
ciliary rootlet coiled-coil, rootletin |
148 |
0.92 |
| chr6_14900560_14901049 | 0.25 |
Foxp2 |
forkhead box P2 |
545 |
0.87 |
| chr10_108360046_108360218 | 0.25 |
Gm23105 |
predicted gene, 23105 |
3533 |
0.25 |
| chr11_16865620_16865819 | 0.25 |
Egfr |
epidermal growth factor receptor |
12431 |
0.2 |
| chr3_60965282_60965682 | 0.25 |
P2ry1 |
purinergic receptor P2Y, G-protein coupled 1 |
37313 |
0.16 |
| chr4_128947381_128947553 | 0.25 |
Gm15904 |
predicted gene 15904 |
11462 |
0.15 |
| chr13_107646164_107646517 | 0.25 |
Gm32090 |
predicted gene, 32090 |
33491 |
0.16 |
| chr3_145118680_145119047 | 0.24 |
Odf2l |
outer dense fiber of sperm tails 2-like |
111 |
0.97 |
| chr5_150572501_150572662 | 0.24 |
Gm43690 |
predicted gene 43690 |
1483 |
0.19 |
| chr13_53001712_53001870 | 0.24 |
Gm33424 |
predicted gene, 33424 |
14411 |
0.16 |
| chr2_11706768_11706947 | 0.24 |
Il15ra |
interleukin 15 receptor, alpha chain |
1009 |
0.47 |
| chr17_8792731_8792886 | 0.24 |
Pde10a |
phosphodiesterase 10A |
8885 |
0.24 |
| chr9_85413933_85414088 | 0.24 |
Gm48833 |
predicted gene, 48833 |
5148 |
0.22 |
| chr15_38519170_38519716 | 0.24 |
Azin1 |
antizyme inhibitor 1 |
177 |
0.84 |
| chr12_104856908_104857124 | 0.23 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
355 |
0.86 |
| chr19_43463068_43463222 | 0.23 |
Gm47938 |
predicted gene, 47938 |
6613 |
0.14 |
| chr4_108085569_108085740 | 0.23 |
Mir6397 |
microRNA 6397 |
1703 |
0.28 |
| chr11_4190861_4191026 | 0.23 |
Tbc1d10a |
TBC1 domain family, member 10a |
4121 |
0.11 |
| chr18_31711636_31711807 | 0.23 |
Gm50060 |
predicted gene, 50060 |
26994 |
0.14 |
| chr3_97226229_97226380 | 0.23 |
Bcl9 |
B cell CLL/lymphoma 9 |
1060 |
0.5 |
| chr10_127936742_127936977 | 0.23 |
Gm15900 |
predicted gene 15900 |
11701 |
0.09 |
| chr5_32713672_32713870 | 0.23 |
Gm43852 |
predicted gene 43852 |
217 |
0.89 |
| chr2_166197123_166197281 | 0.23 |
Gm11466 |
predicted gene 11466 |
6340 |
0.16 |
| chr18_11657291_11657484 | 0.23 |
Rbbp8 |
retinoblastoma binding protein 8, endonuclease |
4 |
0.87 |
| chr2_6165912_6166067 | 0.23 |
A230108P19Rik |
RIKEN cDNA A230108P19 gene |
27262 |
0.12 |
| chr13_95639510_95639665 | 0.23 |
F2r |
coagulation factor II (thrombin) receptor |
21100 |
0.13 |
| chr15_97025909_97026070 | 0.23 |
Slc38a4 |
solute carrier family 38, member 4 |
5222 |
0.3 |
| chr3_118652951_118653308 | 0.23 |
Dpyd |
dihydropyrimidine dehydrogenase |
90943 |
0.08 |
| chr1_74066477_74066680 | 0.23 |
Tns1 |
tensin 1 |
24773 |
0.17 |
| chr2_167777355_167777522 | 0.23 |
Gm14321 |
predicted gene 14321 |
29 |
0.97 |
| chr3_108840435_108840756 | 0.23 |
Stxbp3 |
syntaxin binding protein 3 |
69 |
0.97 |
| chr12_91846606_91846757 | 0.23 |
Sel1l |
sel-1 suppressor of lin-12-like (C. elegans) |
2448 |
0.23 |
| chr11_94044111_94044443 | 0.23 |
Spag9 |
sperm associated antigen 9 |
9 |
0.98 |
| chr1_190919860_190920026 | 0.23 |
Angel2 |
angel homolog 2 |
5169 |
0.17 |
| chr18_33214764_33214929 | 0.22 |
Stard4 |
StAR-related lipid transfer (START) domain containing 4 |
984 |
0.69 |
| chr14_120312913_120313266 | 0.22 |
Mbnl2 |
muscleblind like splicing factor 2 |
6633 |
0.28 |
| chr3_148899467_148899618 | 0.22 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
55093 |
0.14 |
| chrX_170009083_170009242 | 0.22 |
Erdr1 |
erythroid differentiation regulator 1 |
497 |
0.77 |
| chr19_17617854_17618016 | 0.22 |
Gm17819 |
predicted gene, 17819 |
74828 |
0.11 |
| chr17_53478848_53479280 | 0.21 |
Rab5a |
RAB5A, member RAS oncogene family |
170 |
0.93 |
| chr19_10122335_10122508 | 0.21 |
Fads2 |
fatty acid desaturase 2 |
20675 |
0.12 |
| chr14_69629438_69629774 | 0.21 |
Gm27177 |
predicted gene 27177 |
10668 |
0.12 |
| chr11_79254630_79254838 | 0.21 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
63 |
0.97 |
| chr12_54459633_54459834 | 0.21 |
Gm7557 |
predicted gene 7557 |
29335 |
0.13 |
| chr17_34024597_34024949 | 0.21 |
Ring1 |
ring finger protein 1 |
93 |
0.48 |
| chr4_137919760_137920083 | 0.21 |
Ece1 |
endothelin converting enzyme 1 |
6239 |
0.22 |
| chr18_73844340_73844698 | 0.21 |
Mro |
maestro |
14866 |
0.2 |
| chr17_5261920_5262119 | 0.21 |
Gm29050 |
predicted gene 29050 |
126744 |
0.05 |
| chr15_33687773_33687947 | 0.20 |
Tspyl5 |
testis-specific protein, Y-encoded-like 5 |
24 |
0.98 |
| chr14_66103708_66103859 | 0.20 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
6877 |
0.17 |
| chr2_60124236_60125022 | 0.20 |
Gm13620 |
predicted gene 13620 |
467 |
0.65 |
| chr3_21878552_21878734 | 0.20 |
7530428D23Rik |
RIKEN cDNA 7530428D23 gene |
77870 |
0.1 |
| chr15_67153583_67153747 | 0.20 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
23047 |
0.23 |
| chr2_20896513_20896664 | 0.20 |
Arhgap21 |
Rho GTPase activating protein 21 |
13359 |
0.21 |
| chr16_16829274_16829425 | 0.20 |
Spag6l |
sperm associated antigen 6-like |
5 |
0.96 |
| chr6_28981358_28981514 | 0.20 |
Gm3294 |
predicted gene 3294 |
198 |
0.95 |
| chr8_76148011_76148197 | 0.20 |
Gm45742 |
predicted gene 45742 |
31077 |
0.21 |
| chr7_25248454_25248809 | 0.20 |
Erf |
Ets2 repressor factor |
2099 |
0.14 |
| chrY_90799049_90799217 | 0.20 |
Gm47283 |
predicted gene, 47283 |
8682 |
0.18 |
| chr3_27145774_27146219 | 0.20 |
Ect2 |
ect2 oncogene |
1205 |
0.44 |
| chr8_60992984_60993275 | 0.20 |
Nek1 |
NIMA (never in mitosis gene a)-related expressed kinase 1 |
66 |
0.96 |
| chr15_7175278_7175429 | 0.20 |
Lifr |
LIF receptor alpha |
21000 |
0.23 |
| chr13_24948941_24949098 | 0.20 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
5867 |
0.14 |
| chr4_105218216_105218614 | 0.20 |
Plpp3 |
phospholipid phosphatase 3 |
61068 |
0.14 |
| chr3_18151182_18151333 | 0.20 |
Gm23686 |
predicted gene, 23686 |
26368 |
0.2 |
| chr9_96478340_96478816 | 0.19 |
Rnf7 |
ring finger protein 7 |
10 |
0.97 |
| chr11_23140920_23141288 | 0.19 |
1700061J23Rik |
RIKEN cDNA 1700061J23 gene |
11036 |
0.18 |
| chr19_44400352_44400581 | 0.19 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6224 |
0.15 |
| chr1_164543147_164543444 | 0.19 |
D630023O14Rik |
RIKEN cDNA D630023O14 gene |
32160 |
0.13 |
| chr11_90130089_90130271 | 0.19 |
Gm11494 |
predicted gene 11494 |
13444 |
0.19 |
| chr2_58756221_58756376 | 0.19 |
Upp2 |
uridine phosphorylase 2 |
1086 |
0.54 |
| chr12_119070550_119070715 | 0.19 |
Gm18921 |
predicted gene, 18921 |
3202 |
0.35 |
| chr10_3973022_3973187 | 0.19 |
Mthfd1l |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
14 |
0.97 |
| chr16_87128308_87128459 | 0.19 |
Gm32865 |
predicted gene, 32865 |
56 |
0.99 |
| chr8_45975033_45975552 | 0.19 |
1700029J07Rik |
RIKEN cDNA 1700029J07 gene |
40 |
0.74 |
| chr11_16786533_16786734 | 0.19 |
Egfr |
epidermal growth factor receptor |
34403 |
0.15 |
| chr11_119105318_119105491 | 0.19 |
Gm11754 |
predicted gene 11754 |
6021 |
0.15 |
| chr2_31476693_31476999 | 0.19 |
Ass1 |
argininosuccinate synthetase 1 |
6639 |
0.2 |
| chr11_5900664_5900815 | 0.19 |
Gck |
glucokinase |
1348 |
0.27 |
| chr4_108425790_108426145 | 0.19 |
Gpx7 |
glutathione peroxidase 7 |
19006 |
0.1 |
| chr1_92473264_92473470 | 0.19 |
Ndufa10 |
NADH:ubiquinone oxidoreductase subunit A10 |
380 |
0.77 |
| chr5_87574485_87574667 | 0.19 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
5549 |
0.12 |
| chr9_20492670_20492836 | 0.18 |
Zfp426 |
zinc finger protein 426 |
7 |
0.96 |
| chr11_118723319_118723470 | 0.18 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
37647 |
0.17 |
| chr17_24168769_24169218 | 0.18 |
Atp6v0c |
ATPase, H+ transporting, lysosomal V0 subunit C |
73 |
0.93 |
| chr3_129578277_129578428 | 0.18 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
25972 |
0.15 |
| chr2_58794623_58794807 | 0.18 |
Upp2 |
uridine phosphorylase 2 |
29390 |
0.17 |
| chr2_31742282_31742433 | 0.18 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
17586 |
0.14 |
| chr9_43224415_43225053 | 0.18 |
Oaf |
out at first homolog |
358 |
0.84 |
| chr5_125524109_125525122 | 0.18 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr8_69192982_69193338 | 0.18 |
Lzts1 |
leucine zipper, putative tumor suppressor 1 |
8935 |
0.15 |
| chr5_125521313_125521481 | 0.18 |
Aacs |
acetoacetyl-CoA synthetase |
6154 |
0.17 |
| chr10_76997859_76998018 | 0.18 |
Gm35721 |
predicted gene, 35721 |
10631 |
0.12 |
| chr15_101453443_101453620 | 0.18 |
Krt88 |
keratin 88 |
5787 |
0.08 |
| chr10_106941491_106941653 | 0.18 |
Gm19007 |
predicted gene, 19007 |
7725 |
0.25 |
| chr8_122306649_122307232 | 0.18 |
Zfpm1 |
zinc finger protein, multitype 1 |
380 |
0.82 |
| chr16_43464306_43464503 | 0.18 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
39210 |
0.15 |
| chr2_26928748_26928911 | 0.18 |
Surf4 |
surfeit gene 4 |
4554 |
0.08 |
| chr11_101302224_101302524 | 0.18 |
Becn1 |
beclin 1, autophagy related |
88 |
0.9 |
| chr3_31094797_31095132 | 0.17 |
Skil |
SKI-like |
94 |
0.97 |
| chr16_33966968_33967146 | 0.17 |
Umps |
uridine monophosphate synthetase |
19 |
0.94 |
| chr15_68363065_68363728 | 0.17 |
Gm20732 |
predicted gene 20732 |
220 |
0.9 |
| chr2_38368865_38369094 | 0.17 |
Gm13584 |
predicted gene 13584 |
4999 |
0.16 |
| chr13_64152199_64152545 | 0.17 |
Zfp367 |
zinc finger protein 367 |
113 |
0.63 |
| chr9_105792281_105792501 | 0.17 |
Col6a6 |
collagen, type VI, alpha 6 |
17257 |
0.2 |
| chr6_30304499_30304656 | 0.17 |
1700095J07Rik |
RIKEN cDNA 1700095J07 gene |
28 |
0.5 |
| chr3_145924039_145924997 | 0.17 |
Bcl10 |
B cell leukemia/lymphoma 10 |
161 |
0.95 |
| chr2_72160606_72160802 | 0.17 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
11908 |
0.19 |
| chr13_109485029_109485208 | 0.17 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
42935 |
0.21 |
| chr11_16889902_16890060 | 0.17 |
Egfr |
epidermal growth factor receptor |
11831 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 1.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.2 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.1 | 0.2 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 | 0.2 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.3 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.0 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.2 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.0 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.0 | GO:0043465 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0034950 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0008579 | JUN kinase phosphatase activity(GO:0008579) |
| 0.0 | 0.1 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |