Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
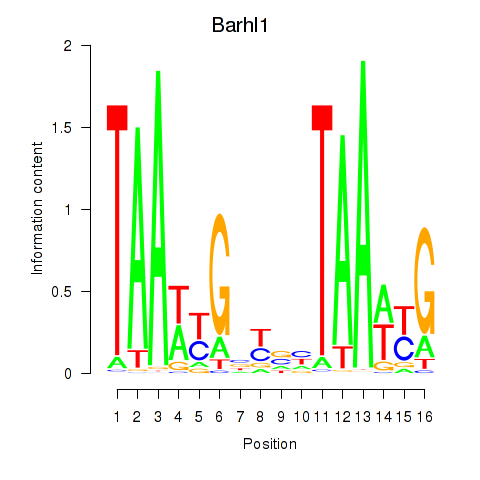
Results for Barhl1
Z-value: 1.63
Transcription factors associated with Barhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl1
|
ENSMUSG00000026805.8 | BarH like homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_28905683_28905848 | Barhl1 | 6730 | 0.175014 | -0.72 | 1.1e-01 | Click! |
| chr2_28909920_28910079 | Barhl1 | 2496 | 0.252866 | -0.66 | 1.5e-01 | Click! |
| chr2_28908189_28908510 | Barhl1 | 4146 | 0.198062 | -0.50 | 3.1e-01 | Click! |
| chr2_28924014_28924175 | Barhl1 | 7426 | 0.174094 | 0.46 | 3.6e-01 | Click! |
| chr2_28916156_28916316 | Barhl1 | 176 | 0.943928 | 0.17 | 7.5e-01 | Click! |
Activity of the Barhl1 motif across conditions
Conditions sorted by the z-value of the Barhl1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_93185394_93185584 | 1.48 |
Gm45909 |
predicted gene 45909 |
5869 |
0.14 |
| chr9_106231818_106232229 | 1.31 |
Alas1 |
aminolevulinic acid synthase 1 |
5061 |
0.11 |
| chr5_29596501_29596658 | 1.01 |
Ube3c |
ubiquitin protein ligase E3C |
5750 |
0.22 |
| chr16_42998647_42998825 | 0.99 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43105 |
0.16 |
| chr3_51228745_51228916 | 0.97 |
Gm38357 |
predicted gene, 38357 |
3087 |
0.18 |
| chr2_50284584_50284735 | 0.94 |
Mmadhc |
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
4540 |
0.25 |
| chr3_51185131_51185304 | 0.87 |
Noct |
nocturnin |
39230 |
0.12 |
| chr16_26697784_26698094 | 0.85 |
Il1rap |
interleukin 1 receptor accessory protein |
24495 |
0.23 |
| chr5_77314693_77314994 | 0.84 |
Gm42758 |
predicted gene 42758 |
3812 |
0.15 |
| chr11_72683212_72683363 | 0.81 |
Ankfy1 |
ankyrin repeat and FYVE domain containing 1 |
6719 |
0.18 |
| chr5_90322868_90323041 | 0.80 |
Ankrd17 |
ankyrin repeat domain 17 |
16789 |
0.19 |
| chr7_131446025_131446176 | 0.77 |
Gm44982 |
predicted gene 44982 |
5629 |
0.11 |
| chr13_48540722_48541023 | 0.77 |
Mirlet7a-1 |
microRNA let7a-1 |
2600 |
0.12 |
| chr6_59453276_59453536 | 0.76 |
Gprin3 |
GPRIN family member 3 |
27112 |
0.26 |
| chr19_21624893_21625081 | 0.73 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
5781 |
0.23 |
| chr4_99046846_99047148 | 0.73 |
Dock7 |
dedicator of cytokinesis 7 |
8336 |
0.18 |
| chr7_136457949_136458342 | 0.71 |
Gm36849 |
predicted gene, 36849 |
104781 |
0.07 |
| chr18_86383968_86384119 | 0.71 |
Neto1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
10909 |
0.24 |
| chr2_42039001_42039161 | 0.70 |
Gm13461 |
predicted gene 13461 |
50252 |
0.18 |
| chr9_94083843_94084083 | 0.68 |
Gm5369 |
predicted gene 5369 |
55499 |
0.17 |
| chr10_111583801_111583952 | 0.68 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
10397 |
0.16 |
| chr1_21299781_21300307 | 0.68 |
Gm4956 |
predicted gene 4956 |
1732 |
0.22 |
| chr3_144446211_144446394 | 0.67 |
Gm5857 |
predicted gene 5857 |
18630 |
0.18 |
| chr2_15064153_15064369 | 0.67 |
Arl5b |
ADP-ribosylation factor-like 5B |
3878 |
0.17 |
| chr17_28428613_28428954 | 0.65 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr13_18729450_18729601 | 0.65 |
Vps41 |
VPS41 HOPS complex subunit |
12235 |
0.27 |
| chr16_26619429_26619580 | 0.65 |
Il1rap |
interleukin 1 receptor accessory protein |
4652 |
0.32 |
| chr7_123406898_123407049 | 0.64 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
1207 |
0.47 |
| chr14_8431164_8431315 | 0.63 |
Gm8416 |
predicted gene 8416 |
22322 |
0.19 |
| chr1_67169083_67169234 | 0.63 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
46132 |
0.15 |
| chr9_55278709_55278860 | 0.62 |
Nrg4 |
neuregulin 4 |
4788 |
0.2 |
| chr19_12793824_12793975 | 0.61 |
Zfp91 |
zinc finger protein 91 |
2227 |
0.17 |
| chr8_36251417_36251586 | 0.60 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
1985 |
0.35 |
| chr1_84261308_84261647 | 0.60 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
22946 |
0.2 |
| chr11_75421874_75422240 | 0.59 |
Serpinf1 |
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
495 |
0.56 |
| chr8_83590177_83590330 | 0.59 |
Tecr |
trans-2,3-enoyl-CoA reductase |
4168 |
0.11 |
| chr10_87936414_87936806 | 0.59 |
Tyms-ps |
thymidylate synthase, pseudogene |
30237 |
0.15 |
| chr12_104041015_104041169 | 0.58 |
Serpina12 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
3351 |
0.13 |
| chr12_8081681_8081832 | 0.58 |
Apob |
apolipoprotein B |
69397 |
0.1 |
| chrX_36228961_36229143 | 0.57 |
Gm6023 |
predicted gene 6023 |
14923 |
0.15 |
| chr2_22670750_22670915 | 0.57 |
Gm23636 |
predicted gene, 23636 |
4174 |
0.17 |
| chr7_13637528_13637679 | 0.57 |
Zbed4-ps1 |
zinc finger, BED type containing 4, pseudogene 1 |
1504 |
0.32 |
| chr7_131362078_131362229 | 0.57 |
2310057M21Rik |
RIKEN cDNA 2310057M21 gene |
234 |
0.85 |
| chr5_30818633_30818850 | 0.57 |
Gm19409 |
predicted gene, 19409 |
3121 |
0.13 |
| chr7_52055664_52055818 | 0.56 |
Gm22211 |
predicted gene, 22211 |
19838 |
0.16 |
| chr14_75309433_75309600 | 0.56 |
Zc3h13 |
zinc finger CCCH type containing 13 |
25130 |
0.14 |
| chr10_87869244_87869395 | 0.56 |
Igf1os |
insulin-like growth factor 1, opposite strand |
5938 |
0.21 |
| chr7_91514549_91514720 | 0.56 |
Gm44679 |
predicted gene 44679 |
4481 |
0.21 |
| chr4_48244997_48245148 | 0.55 |
Erp44 |
endoplasmic reticulum protein 44 |
598 |
0.76 |
| chr5_52147484_52147676 | 0.55 |
Gm43176 |
predicted gene 43176 |
5007 |
0.13 |
| chr9_57065617_57065768 | 0.55 |
Gm10658 |
predicted gene 10658 |
6115 |
0.13 |
| chr14_21287151_21287302 | 0.54 |
Adk |
adenosine kinase |
30899 |
0.22 |
| chr16_26727363_26727514 | 0.54 |
Il1rap |
interleukin 1 receptor accessory protein |
396 |
0.9 |
| chr12_86516484_86516661 | 0.54 |
Esrrb |
estrogen related receptor, beta |
12163 |
0.24 |
| chr3_5346748_5346907 | 0.54 |
Zfhx4 |
zinc finger homeodomain 4 |
105155 |
0.07 |
| chr16_46002412_46002585 | 0.54 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
7720 |
0.17 |
| chr2_147949307_147949458 | 0.54 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
26803 |
0.2 |
| chr14_101841283_101841436 | 0.53 |
Lmo7 |
LIM domain only 7 |
540 |
0.85 |
| chr15_95744172_95744337 | 0.53 |
Gm6961 |
predicted gene 6961 |
7337 |
0.13 |
| chr1_67208135_67208300 | 0.52 |
Gm15668 |
predicted gene 15668 |
40983 |
0.16 |
| chr1_67175551_67175991 | 0.52 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
52745 |
0.13 |
| chr19_20512305_20512469 | 0.52 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
19627 |
0.19 |
| chr9_80335524_80335697 | 0.52 |
Myo6 |
myosin VI |
28696 |
0.2 |
| chr2_166974855_166975035 | 0.52 |
Stau1 |
staufen double-stranded RNA binding protein 1 |
1826 |
0.25 |
| chr9_14381410_14381579 | 0.52 |
Endod1 |
endonuclease domain containing 1 |
13 |
0.96 |
| chr5_92607049_92607221 | 0.51 |
Stbd1 |
starch binding domain 1 |
4067 |
0.19 |
| chr2_114788136_114788290 | 0.51 |
Gm13974 |
predicted gene 13974 |
65880 |
0.12 |
| chr3_98697303_98697632 | 0.51 |
Gm12400 |
predicted gene 12400 |
9285 |
0.13 |
| chr10_25127208_25127439 | 0.49 |
Akap7 |
A kinase (PRKA) anchor protein 7 |
72835 |
0.08 |
| chr4_109867828_109867979 | 0.49 |
Faf1 |
Fas-associated factor 1 |
27118 |
0.21 |
| chr10_53929773_53930076 | 0.49 |
Man1a |
mannosidase 1, alpha |
47191 |
0.16 |
| chr6_70874912_70875301 | 0.49 |
Eif2ak3 |
eukaryotic translation initiation factor 2 alpha kinase 3 |
3455 |
0.18 |
| chr14_64695745_64695916 | 0.49 |
Kif13b |
kinesin family member 13B |
43228 |
0.13 |
| chr5_90891820_90892079 | 0.49 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
706 |
0.49 |
| chrX_77496822_77496989 | 0.48 |
Tbl1x |
transducin (beta)-like 1 X-linked |
14108 |
0.22 |
| chr8_36250043_36250194 | 0.48 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
602 |
0.76 |
| chr5_123091308_123091459 | 0.48 |
Tmem120b |
transmembrane protein 120B |
8518 |
0.09 |
| chr19_39740815_39740983 | 0.48 |
Cyp2c68 |
cytochrome P450, family 2, subfamily c, polypeptide 68 |
155 |
0.97 |
| chr5_147727096_147727252 | 0.48 |
Flt1 |
FMS-like tyrosine kinase 1 |
1163 |
0.52 |
| chr4_40145230_40145427 | 0.48 |
Aco1 |
aconitase 1 |
2247 |
0.3 |
| chr15_62746422_62746596 | 0.48 |
Gm22521 |
predicted gene, 22521 |
46692 |
0.18 |
| chr3_51204305_51204466 | 0.48 |
Noct |
nocturnin |
20062 |
0.14 |
| chr7_64347926_64348121 | 0.47 |
Gm20457 |
predicted gene 20457 |
2059 |
0.24 |
| chr4_48275903_48276069 | 0.47 |
Erp44 |
endoplasmic reticulum protein 44 |
3466 |
0.23 |
| chr7_19508824_19509003 | 0.47 |
Trappc6a |
trafficking protein particle complex 6A |
183 |
0.76 |
| chr1_21328678_21328866 | 0.47 |
Gm21909 |
predicted gene, 21909 |
12452 |
0.09 |
| chr6_58651386_58651542 | 0.47 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
10882 |
0.23 |
| chr9_65013148_65013299 | 0.47 |
Hacd3 |
3-hydroxyacyl-CoA dehydratase 3 |
8470 |
0.14 |
| chr8_125012381_125013106 | 0.47 |
Tsnax |
translin-associated factor X |
254 |
0.9 |
| chr14_61687453_61687640 | 0.47 |
Gm37820 |
predicted gene, 37820 |
4036 |
0.12 |
| chr2_174774160_174774316 | 0.47 |
Edn3 |
endothelin 3 |
4316 |
0.25 |
| chr4_125850425_125850576 | 0.46 |
2610028E06Rik |
RIKEN cDNA 2610028E06 gene |
48482 |
0.16 |
| chr18_75371968_75372846 | 0.46 |
Smad7 |
SMAD family member 7 |
2431 |
0.3 |
| chr8_35411428_35411600 | 0.46 |
Gm45301 |
predicted gene 45301 |
2008 |
0.29 |
| chr5_121719683_121720110 | 0.46 |
Gm43571 |
predicted gene 43571 |
4787 |
0.12 |
| chr6_54856005_54856204 | 0.46 |
Znrf2 |
zinc and ring finger 2 |
38656 |
0.14 |
| chr5_105394182_105394457 | 0.46 |
Gm26519 |
predicted gene, 26519 |
6926 |
0.16 |
| chr18_31704198_31704968 | 0.46 |
Gm50060 |
predicted gene, 50060 |
34132 |
0.13 |
| chr4_60089071_60089222 | 0.45 |
Mup-ps1 |
major urinary protein, pseudogene 1 |
3306 |
0.22 |
| chr7_113268439_113268597 | 0.45 |
Gm45355 |
predicted gene 45355 |
26819 |
0.15 |
| chr4_150756055_150756229 | 0.45 |
Gm13049 |
predicted gene 13049 |
69591 |
0.08 |
| chr7_27120303_27120454 | 0.45 |
Cyp2f2 |
cytochrome P450, family 2, subfamily f, polypeptide 2 |
417 |
0.69 |
| chr4_143194960_143195165 | 0.45 |
Prdm2 |
PR domain containing 2, with ZNF domain |
327 |
0.85 |
| chr6_22352199_22352350 | 0.45 |
Fam3c |
family with sequence similarity 3, member C |
3799 |
0.32 |
| chr6_28524838_28524989 | 0.45 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
36904 |
0.11 |
| chr3_149322002_149322153 | 0.45 |
Gm30382 |
predicted gene, 30382 |
1797 |
0.35 |
| chr17_28707641_28707845 | 0.45 |
Mapk14 |
mitogen-activated protein kinase 14 |
15175 |
0.11 |
| chr11_104441052_104441411 | 0.44 |
Kansl1 |
KAT8 regulatory NSL complex subunit 1 |
471 |
0.79 |
| chr4_14814387_14814544 | 0.44 |
Otud6b |
OTU domain containing 6B |
11930 |
0.18 |
| chr16_38097146_38097299 | 0.43 |
Gsk3b |
glycogen synthase kinase 3 beta |
6934 |
0.23 |
| chr11_60877088_60877239 | 0.43 |
Tmem11 |
transmembrane protein 11 |
1624 |
0.26 |
| chr7_81575164_81575561 | 0.43 |
Whamm |
WAS protein homolog associated with actin, golgi membranes and microtubules |
3418 |
0.13 |
| chr12_84201804_84201955 | 0.43 |
Gm31513 |
predicted gene, 31513 |
5910 |
0.11 |
| chr13_4041060_4041239 | 0.43 |
Gm18857 |
predicted gene, 18857 |
5904 |
0.14 |
| chr16_14707577_14707794 | 0.42 |
Snai2 |
snail family zinc finger 2 |
1833 |
0.43 |
| chr2_110304106_110304292 | 0.42 |
Bbox1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
1542 |
0.45 |
| chr8_93199495_93199658 | 0.42 |
Ces1d |
carboxylesterase 1D |
1738 |
0.27 |
| chr6_31253045_31253249 | 0.41 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
24701 |
0.14 |
| chr14_20931681_20931890 | 0.41 |
Vcl |
vinculin |
2387 |
0.32 |
| chr1_160781397_160781614 | 0.41 |
Rabgap1l |
RAB GTPase activating protein 1-like |
11433 |
0.12 |
| chr18_75449947_75450120 | 0.41 |
Gm10532 |
predicted gene 10532 |
64612 |
0.11 |
| chr11_28702228_28702389 | 0.41 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
20744 |
0.15 |
| chr8_40903804_40903955 | 0.41 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
5441 |
0.18 |
| chr5_25154949_25155325 | 0.41 |
Gm26051 |
predicted gene, 26051 |
13327 |
0.14 |
| chr19_60788929_60789115 | 0.41 |
Eif3a |
eukaryotic translation initiation factor 3, subunit A |
1621 |
0.24 |
| chr19_40174897_40175092 | 0.40 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
12292 |
0.15 |
| chr1_67182575_67183305 | 0.40 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59914 |
0.11 |
| chr7_49459741_49459913 | 0.40 |
Nav2 |
neuron navigator 2 |
9067 |
0.23 |
| chr3_22077324_22077923 | 0.40 |
Tbl1xr1 |
transducin (beta)-like 1X-linked receptor 1 |
339 |
0.82 |
| chr18_12668617_12668967 | 0.40 |
Gm41668 |
predicted gene, 41668 |
20373 |
0.13 |
| chr18_16798014_16798260 | 0.40 |
Cdh2 |
cadherin 2 |
10325 |
0.2 |
| chr18_17623971_17624154 | 0.40 |
4930545E07Rik |
RIKEN cDNA 4930545E07 gene |
43072 |
0.18 |
| chr3_129380919_129381082 | 0.40 |
Gm42630 |
predicted gene 42630 |
6890 |
0.17 |
| chr15_95749618_95750040 | 0.40 |
Gm6961 |
predicted gene 6961 |
1762 |
0.25 |
| chr8_93077902_93078174 | 0.40 |
Ces1b |
carboxylesterase 1B |
1979 |
0.28 |
| chr6_24348537_24348787 | 0.40 |
Iqub |
IQ motif and ubiquitin domain containing |
166405 |
0.03 |
| chr15_38058115_38058382 | 0.40 |
Gm22353 |
predicted gene, 22353 |
37 |
0.97 |
| chr4_104893870_104894082 | 0.40 |
C8a |
complement component 8, alpha polypeptide |
17578 |
0.18 |
| chr2_115450732_115450919 | 0.40 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
61376 |
0.12 |
| chr17_74273467_74273665 | 0.39 |
Memo1 |
mediator of cell motility 1 |
15082 |
0.13 |
| chr4_75259626_75259802 | 0.39 |
Dmac1 |
distal membrane arm assembly complex 1 |
18591 |
0.19 |
| chr13_75343113_75343287 | 0.39 |
Gm48234 |
predicted gene, 48234 |
136187 |
0.04 |
| chr16_13257585_13257940 | 0.39 |
Mrtfb |
myocardin related transcription factor B |
1256 |
0.58 |
| chr6_47799355_47799532 | 0.39 |
Mir704 |
microRNA 704 |
4209 |
0.13 |
| chr3_118494272_118494441 | 0.39 |
Gm37773 |
predicted gene, 37773 |
31795 |
0.11 |
| chr11_90675070_90675221 | 0.39 |
Tom1l1 |
target of myb1-like 1 (chicken) |
11833 |
0.2 |
| chr4_71658237_71658395 | 0.39 |
Gm11231 |
predicted gene 11231 |
19697 |
0.27 |
| chr19_59343091_59343283 | 0.39 |
Pdzd8 |
PDZ domain containing 8 |
2593 |
0.21 |
| chr7_132953914_132954065 | 0.39 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
4405 |
0.17 |
| chr13_114485155_114485339 | 0.39 |
Fst |
follistatin |
26296 |
0.14 |
| chr2_67881265_67881416 | 0.39 |
Gm37964 |
predicted gene, 37964 |
17256 |
0.24 |
| chr19_29520625_29520778 | 0.38 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
269 |
0.89 |
| chr4_43393425_43393582 | 0.38 |
Rusc2 |
RUN and SH3 domain containing 2 |
7735 |
0.14 |
| chr7_26313878_26314034 | 0.38 |
Gm42375 |
predicted gene, 42375 |
6670 |
0.14 |
| chr10_95949807_95950050 | 0.38 |
Eea1 |
early endosome antigen 1 |
9171 |
0.18 |
| chr2_78656286_78656827 | 0.38 |
Gm14463 |
predicted gene 14463 |
873 |
0.71 |
| chr6_149169218_149169449 | 0.38 |
Amn1 |
antagonist of mitotic exit network 1 |
4038 |
0.17 |
| chr15_27461169_27461365 | 0.38 |
Gm36899 |
predicted gene, 36899 |
2182 |
0.26 |
| chr8_68061539_68061690 | 0.38 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
613 |
0.81 |
| chr4_76190772_76190944 | 0.38 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
56715 |
0.17 |
| chr16_43234125_43234650 | 0.38 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1493 |
0.45 |
| chr11_95454483_95454634 | 0.37 |
Gm11522 |
predicted gene 11522 |
8163 |
0.15 |
| chr4_6225587_6225787 | 0.37 |
Ubxn2b |
UBX domain protein 2B |
34589 |
0.15 |
| chr15_53166243_53166634 | 0.37 |
Ext1 |
exostosin glycosyltransferase 1 |
29817 |
0.26 |
| chr11_76146489_76146899 | 0.37 |
Gm12340 |
predicted gene 12340 |
2436 |
0.24 |
| chr6_129471487_129471654 | 0.37 |
Clec7a |
C-type lectin domain family 7, member a |
1202 |
0.3 |
| chr15_3469696_3469847 | 0.37 |
Ghr |
growth hormone receptor |
1873 |
0.48 |
| chr4_33033993_33034314 | 0.37 |
Ube2j1 |
ubiquitin-conjugating enzyme E2J 1 |
2575 |
0.17 |
| chr3_79849029_79849198 | 0.37 |
Tmem144 |
transmembrane protein 144 |
3655 |
0.21 |
| chr7_144776184_144776340 | 0.37 |
Gm44929 |
predicted gene 44929 |
11348 |
0.11 |
| chr13_112473176_112473327 | 0.37 |
Il6st |
interleukin 6 signal transducer |
652 |
0.68 |
| chr2_148026747_148026997 | 0.37 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
11398 |
0.17 |
| chr3_119638719_119638870 | 0.36 |
Gm23432 |
predicted gene, 23432 |
80 |
0.98 |
| chr9_70240760_70241034 | 0.36 |
Myo1e |
myosin IE |
33529 |
0.17 |
| chr6_72600544_72600754 | 0.36 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
2021 |
0.15 |
| chr9_53308258_53308435 | 0.36 |
Exph5 |
exophilin 5 |
6676 |
0.2 |
| chr10_88505415_88505608 | 0.36 |
Chpt1 |
choline phosphotransferase 1 |
1438 |
0.36 |
| chr3_121867071_121867223 | 0.36 |
Gm42593 |
predicted gene 42593 |
4305 |
0.2 |
| chr1_44576462_44576616 | 0.35 |
Gm37626 |
predicted gene, 37626 |
15654 |
0.19 |
| chr19_32603829_32604061 | 0.35 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
8155 |
0.23 |
| chrX_100480103_100480280 | 0.35 |
Igbp1 |
immunoglobulin (CD79A) binding protein 1 |
14100 |
0.15 |
| chr6_144201120_144201290 | 0.35 |
Sox5 |
SRY (sex determining region Y)-box 5 |
2852 |
0.41 |
| chr6_52673820_52673989 | 0.35 |
Hibadh |
3-hydroxyisobutyrate dehydrogenase |
33515 |
0.11 |
| chr4_122982836_122983021 | 0.35 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
12724 |
0.13 |
| chr4_76448167_76448318 | 0.35 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
1738 |
0.38 |
| chr16_26688559_26688783 | 0.35 |
Il1rap |
interleukin 1 receptor accessory protein |
33763 |
0.21 |
| chr2_110303618_110304067 | 0.35 |
Bbox1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
1899 |
0.39 |
| chr13_98956021_98956191 | 0.35 |
Gm35215 |
predicted gene, 35215 |
10190 |
0.12 |
| chr14_19986275_19986426 | 0.35 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
8723 |
0.21 |
| chr11_111996214_111996404 | 0.35 |
Gm11679 |
predicted gene 11679 |
47269 |
0.19 |
| chr6_149147053_149147326 | 0.34 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
5543 |
0.14 |
| chr10_20486313_20486464 | 0.34 |
Gm17229 |
predicted gene 17229 |
31776 |
0.16 |
| chr4_80924658_80924891 | 0.34 |
Lurap1l |
leucine rich adaptor protein 1-like |
14128 |
0.24 |
| chr4_10400904_10401065 | 0.34 |
Gm11814 |
predicted gene 11814 |
38759 |
0.2 |
| chr6_22351338_22351531 | 0.34 |
Fam3c |
family with sequence similarity 3, member C |
4639 |
0.3 |
| chr6_108666051_108666462 | 0.34 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
3210 |
0.21 |
| chr3_84285532_84285981 | 0.34 |
Trim2 |
tripartite motif-containing 2 |
14965 |
0.24 |
| chr16_42998828_42999221 | 0.34 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43393 |
0.16 |
| chr2_85089101_85089275 | 0.34 |
4930443O20Rik |
RIKEN cDNA 4930443O20 gene |
2727 |
0.18 |
| chr16_65617781_65617932 | 0.34 |
Gm49633 |
predicted gene, 49633 |
4437 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.4 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.4 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.1 | 0.2 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 0.2 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.0 | 0.0 | GO:0060768 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.0 | 0.3 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.0 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:1901203 | positive regulation of extracellular matrix assembly(GO:1901203) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.0 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.2 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0045738 | negative regulation of DNA repair(GO:0045738) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.4 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.0 | GO:0003140 | determination of left/right asymmetry in lateral mesoderm(GO:0003140) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 1.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0018597 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.3 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.0 | GO:0043907 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0044688 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.0 | GO:0018423 | protein C-terminal leucine carboxyl O-methyltransferase activity(GO:0018423) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |