Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
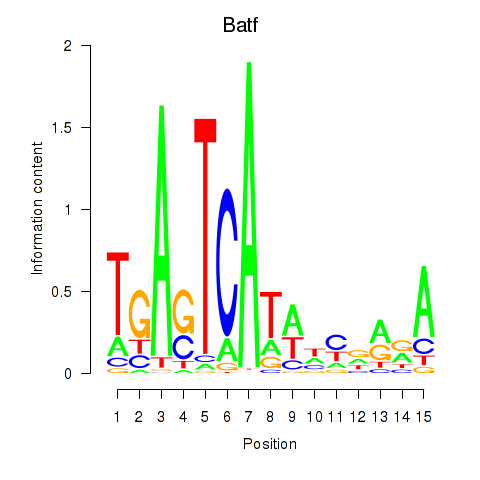
Results for Batf
Z-value: 1.35
Transcription factors associated with Batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf
|
ENSMUSG00000034266.5 | basic leucine zipper transcription factor, ATF-like |
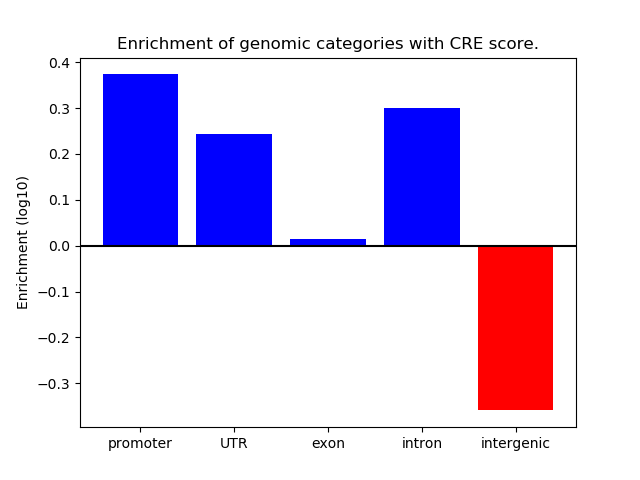
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_85698292_85698454 | Batf | 9081 | 0.128209 | -0.10 | 8.5e-01 | Click! |
Activity of the Batf motif across conditions
Conditions sorted by the z-value of the Batf motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
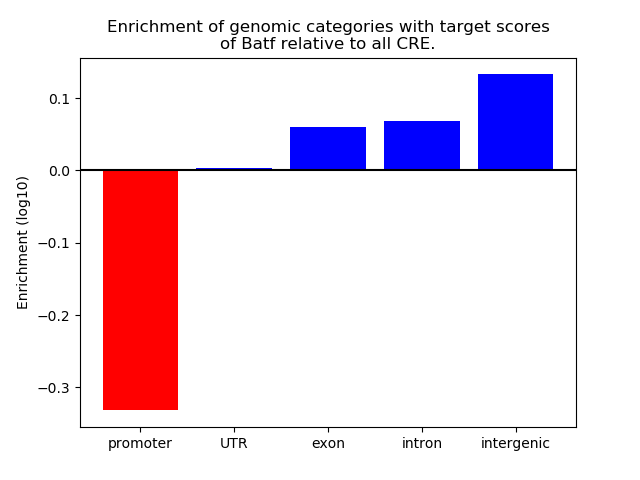
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_46997805_46997956 | 0.87 |
Gm22791 |
predicted gene, 22791 |
12399 |
0.15 |
| chr18_46998104_46998255 | 0.66 |
Gm22791 |
predicted gene, 22791 |
12698 |
0.15 |
| chr5_21384627_21384845 | 0.57 |
Fgl2 |
fibrinogen-like protein 2 |
12094 |
0.18 |
| chr13_44306229_44306411 | 0.49 |
Gm29676 |
predicted gene, 29676 |
25871 |
0.17 |
| chr6_142451928_142452079 | 0.47 |
Gys2 |
glycogen synthase 2 |
21106 |
0.15 |
| chr7_132572852_132573003 | 0.44 |
Oat |
ornithine aminotransferase |
2791 |
0.22 |
| chr6_117604955_117605122 | 0.43 |
Gm45083 |
predicted gene 45083 |
7532 |
0.22 |
| chr15_59448754_59448952 | 0.40 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
74566 |
0.09 |
| chr4_141955631_141955810 | 0.39 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1526 |
0.3 |
| chr5_31036629_31036808 | 0.39 |
Slc5a6 |
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
1276 |
0.28 |
| chr19_32601214_32601377 | 0.39 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
5505 |
0.24 |
| chr1_154077652_154077830 | 0.38 |
Gm25336 |
predicted gene, 25336 |
2458 |
0.27 |
| chr10_98666150_98666367 | 0.36 |
Gm5427 |
predicted gene 5427 |
33452 |
0.22 |
| chr13_54323551_54323702 | 0.36 |
Gm48622 |
predicted gene, 48622 |
4254 |
0.19 |
| chr14_38629749_38629921 | 0.35 |
Gm20641 |
predicted gene 20641 |
107786 |
0.08 |
| chr8_33950546_33950701 | 0.35 |
Rbpms |
RNA binding protein gene with multiple splicing |
20760 |
0.14 |
| chr18_20157783_20157946 | 0.35 |
Dsc1 |
desmocollin 1 |
42993 |
0.17 |
| chr9_65084518_65084694 | 0.35 |
Dpp8 |
dipeptidylpeptidase 8 |
8858 |
0.14 |
| chr15_53163238_53163412 | 0.33 |
Ext1 |
exostosin glycosyltransferase 1 |
32930 |
0.25 |
| chrX_60362413_60362564 | 0.33 |
Mir505 |
microRNA 505 |
32004 |
0.17 |
| chr8_77128069_77128235 | 0.33 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
139 |
0.97 |
| chr11_117914869_117915020 | 0.33 |
Gm11726 |
predicted gene 11726 |
6961 |
0.13 |
| chr6_67441579_67441755 | 0.33 |
Gm44083 |
predicted gene, 44083 |
23616 |
0.11 |
| chr18_34864196_34864456 | 0.33 |
Egr1 |
early growth response 1 |
3119 |
0.18 |
| chr9_84977601_84977752 | 0.32 |
Gm8228 |
predicted gene 8228 |
1161 |
0.55 |
| chr4_57312348_57312552 | 0.32 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
10613 |
0.17 |
| chr11_16737408_16737579 | 0.32 |
Gm25698 |
predicted gene, 25698 |
4782 |
0.22 |
| chr16_8659432_8659599 | 0.31 |
Pmm2 |
phosphomannomutase 2 |
10923 |
0.1 |
| chr11_16860515_16860728 | 0.31 |
Egfr |
epidermal growth factor receptor |
17529 |
0.19 |
| chr7_26313878_26314034 | 0.30 |
Gm42375 |
predicted gene, 42375 |
6670 |
0.14 |
| chr11_16703044_16703296 | 0.30 |
Gm25698 |
predicted gene, 25698 |
29541 |
0.15 |
| chr3_18251077_18251277 | 0.30 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
7839 |
0.24 |
| chr12_28699054_28699358 | 0.30 |
Trappc12 |
trafficking protein particle complex 12 |
2403 |
0.22 |
| chr6_51709095_51709246 | 0.30 |
Gm38811 |
predicted gene, 38811 |
1911 |
0.43 |
| chr19_40158328_40158479 | 0.29 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
28883 |
0.13 |
| chr9_15297335_15297517 | 0.29 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
4088 |
0.06 |
| chr17_70746763_70747075 | 0.29 |
5031415H12Rik |
RIKEN cDNA 5031415H12 gene |
8663 |
0.19 |
| chr2_4542651_4542802 | 0.29 |
Frmd4a |
FERM domain containing 4A |
17028 |
0.2 |
| chr4_105557523_105557702 | 0.29 |
Gm12726 |
predicted gene 12726 |
20728 |
0.24 |
| chr15_7297709_7298002 | 0.29 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
43087 |
0.16 |
| chr2_39161075_39161269 | 0.28 |
Gm16523 |
predicted gene, 16523 |
817 |
0.5 |
| chr1_162987339_162987541 | 0.28 |
Fmo3 |
flavin containing monooxygenase 3 |
2912 |
0.22 |
| chr2_42039219_42039481 | 0.28 |
Gm13461 |
predicted gene 13461 |
50521 |
0.18 |
| chr3_109255070_109255263 | 0.28 |
Gm13865 |
predicted gene 13865 |
62299 |
0.11 |
| chr15_43701477_43701832 | 0.28 |
Gm2140 |
predicted gene 2140 |
106807 |
0.07 |
| chr6_55250906_55251062 | 0.27 |
Mindy4 |
MINDY lysine 48 deubiquitinase 4 |
27171 |
0.15 |
| chr7_51868214_51868365 | 0.27 |
Gas2 |
growth arrest specific 2 |
4385 |
0.17 |
| chr8_56443820_56444020 | 0.27 |
Gm7419 |
predicted gene 7419 |
9595 |
0.2 |
| chr17_25756148_25756299 | 0.26 |
Msln |
mesothelin |
1845 |
0.11 |
| chr14_57107820_57108112 | 0.25 |
Gjb2 |
gap junction protein, beta 2 |
3264 |
0.19 |
| chr2_147988721_147988917 | 0.25 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
12634 |
0.21 |
| chr1_182383191_182383342 | 0.25 |
Gm5706 |
predicted gene 5706 |
10269 |
0.15 |
| chr18_39816253_39816404 | 0.25 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
42831 |
0.17 |
| chr11_60356089_60356253 | 0.25 |
Drc3 |
dynein regulatory complex subunit 3 |
2791 |
0.16 |
| chr7_130968693_130968854 | 0.25 |
Htra1 |
HtrA serine peptidase 1 |
12897 |
0.19 |
| chr6_122654614_122654765 | 0.25 |
Dppa3 |
developmental pluripotency-associated 3 |
27936 |
0.1 |
| chr8_128648206_128648359 | 0.24 |
Gm26002 |
predicted gene, 26002 |
2715 |
0.24 |
| chr13_75789506_75789657 | 0.24 |
Gm8288 |
predicted gene 8288 |
13125 |
0.13 |
| chr1_71723871_71724224 | 0.24 |
Gm5256 |
predicted gene 5256 |
9123 |
0.19 |
| chr8_75372121_75372290 | 0.24 |
Umpk-ps |
uridine monophosphate kinase, pseudogene |
11597 |
0.15 |
| chr7_110953364_110953689 | 0.24 |
Mrvi1 |
MRV integration site 1 |
7339 |
0.2 |
| chr11_4195737_4195910 | 0.24 |
Tbc1d10a |
TBC1 domain family, member 10a |
9001 |
0.1 |
| chrX_74243813_74243964 | 0.24 |
Flna |
filamin, alpha |
2476 |
0.11 |
| chr4_105149185_105149336 | 0.23 |
Plpp3 |
phospholipid phosphatase 3 |
8087 |
0.26 |
| chr13_30036706_30036863 | 0.23 |
Gm11368 |
predicted gene 11368 |
21893 |
0.17 |
| chr13_98317974_98318142 | 0.23 |
Gm9828 |
predicted gene 9828 |
648 |
0.51 |
| chr9_56497408_56497559 | 0.23 |
Gm47175 |
predicted gene, 47175 |
9178 |
0.16 |
| chr7_25411234_25411387 | 0.23 |
Cxcl17 |
chemokine (C-X-C motif) ligand 17 |
1576 |
0.21 |
| chr1_71807937_71808302 | 0.22 |
Gm37217 |
predicted gene, 37217 |
38491 |
0.15 |
| chr8_8947104_8947393 | 0.22 |
Gm44515 |
predicted gene 44515 |
14549 |
0.25 |
| chr2_8116897_8117048 | 0.22 |
Gm13254 |
predicted gene 13254 |
30893 |
0.26 |
| chr3_151298420_151298623 | 0.22 |
4930555A03Rik |
RIKEN cDNA 4930555A03 gene |
3518 |
0.3 |
| chr15_53184591_53184792 | 0.22 |
Ext1 |
exostosin glycosyltransferase 1 |
11564 |
0.31 |
| chr4_133680298_133680626 | 0.22 |
Pigv |
phosphatidylinositol glycan anchor biosynthesis, class V |
7815 |
0.13 |
| chr11_31747063_31747263 | 0.22 |
Gm38061 |
predicted gene, 38061 |
48515 |
0.13 |
| chr4_120978377_120978528 | 0.22 |
Smap2 |
small ArfGAP 2 |
2744 |
0.18 |
| chr19_14736673_14736829 | 0.22 |
Gm26026 |
predicted gene, 26026 |
18271 |
0.26 |
| chr4_49435274_49435479 | 0.22 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
15758 |
0.12 |
| chr2_120518757_120518908 | 0.22 |
Zfp106 |
zinc finger protein 106 |
1714 |
0.3 |
| chr15_102014961_102015124 | 0.22 |
Krt8 |
keratin 8 |
10560 |
0.11 |
| chr15_3644705_3644883 | 0.22 |
Gm22031 |
predicted gene, 22031 |
41397 |
0.17 |
| chr7_75618027_75618186 | 0.22 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
3418 |
0.26 |
| chr6_142708472_142708687 | 0.22 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
6264 |
0.22 |
| chr7_4170342_4170493 | 0.22 |
Cdc42ep5 |
CDC42 effector protein (Rho GTPase binding) 5 |
5557 |
0.09 |
| chr18_34927551_34927712 | 0.21 |
Etf1 |
eukaryotic translation termination factor 1 |
4376 |
0.14 |
| chr17_34143075_34143349 | 0.21 |
H2-DMb2 |
histocompatibility 2, class II, locus Mb2 |
95 |
0.89 |
| chr12_79600452_79600623 | 0.21 |
Rad51b |
RAD51 paralog B |
273184 |
0.01 |
| chr8_116344914_116345106 | 0.21 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
12051 |
0.29 |
| chr10_109182870_109183021 | 0.21 |
Gm44495 |
predicted gene, 44495 |
143906 |
0.04 |
| chr7_92640507_92640658 | 0.21 |
6430511E19Rik |
RIKEN cDNA 6430511E19 gene |
633 |
0.44 |
| chr9_107654301_107654478 | 0.21 |
Slc38a3 |
solute carrier family 38, member 3 |
1631 |
0.17 |
| chr9_116136919_116137123 | 0.21 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38244 |
0.15 |
| chr10_127544019_127544184 | 0.21 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
893 |
0.41 |
| chrX_129698555_129698713 | 0.21 |
Diaph2 |
diaphanous related formin 2 |
51108 |
0.17 |
| chr1_60833760_60833934 | 0.21 |
Gm11581 |
predicted gene 11581 |
24731 |
0.11 |
| chr7_98580551_98580702 | 0.21 |
Emsy |
EMSY, BRCA2-interacting transcriptional repressor |
13609 |
0.15 |
| chr14_71251214_71251453 | 0.21 |
Gm4251 |
predicted gene 4251 |
49 |
0.99 |
| chr2_167293432_167293597 | 0.21 |
Gm11473 |
predicted gene 11473 |
11737 |
0.16 |
| chr7_79308613_79308815 | 0.21 |
Gm39041 |
predicted gene, 39041 |
9689 |
0.14 |
| chr15_77412067_77412222 | 0.21 |
Apol9a |
apolipoprotein L 9a |
1009 |
0.35 |
| chr19_6390399_6390550 | 0.21 |
Pygm |
muscle glycogen phosphorylase |
2237 |
0.14 |
| chr17_34593277_34593428 | 0.21 |
Pbx2 |
pre B cell leukemia homeobox 2 |
43 |
0.91 |
| chr5_51876051_51876244 | 0.20 |
Gm42616 |
predicted gene 42616 |
3455 |
0.22 |
| chr10_33814374_33814533 | 0.20 |
Gm10327 |
predicted pseudogene 10327 |
4955 |
0.17 |
| chr4_9657208_9657376 | 0.20 |
Asph |
aspartate-beta-hydroxylase |
11794 |
0.19 |
| chr14_86301775_86301947 | 0.20 |
Gm32815 |
predicted gene, 32815 |
2127 |
0.23 |
| chr14_51106687_51106910 | 0.20 |
Eddm3b |
epididymal protein 3B |
7628 |
0.08 |
| chr10_87936931_87937082 | 0.20 |
Tyms-ps |
thymidylate synthase, pseudogene |
29841 |
0.15 |
| chrX_169863890_169864041 | 0.20 |
Mid1 |
midline 1 |
15657 |
0.24 |
| chr12_62877016_62877197 | 0.20 |
Gm7763 |
predicted gene 7763 |
6176 |
0.25 |
| chr12_25784515_25784666 | 0.20 |
Gm9321 |
predicted gene 9321 |
35951 |
0.2 |
| chr11_16904746_16904937 | 0.20 |
Egfr |
epidermal growth factor receptor |
344 |
0.88 |
| chr8_40651238_40651389 | 0.20 |
Mtmr7 |
myotubularin related protein 7 |
16516 |
0.14 |
| chr15_67780972_67781123 | 0.20 |
Gm49408 |
predicted gene, 49408 |
92781 |
0.09 |
| chr3_95267928_95268183 | 0.20 |
Prune1 |
prune exopolyphosphatase |
889 |
0.34 |
| chr9_74366017_74366190 | 0.20 |
Nr1h2-ps |
nuclear receptor subfamily 1, group H, member 2, pseudogene |
1040 |
0.56 |
| chr3_132565872_132566040 | 0.20 |
Gm25307 |
predicted gene, 25307 |
28291 |
0.18 |
| chr7_49396252_49396429 | 0.20 |
Nav2 |
neuron navigator 2 |
31619 |
0.18 |
| chr18_38780310_38780655 | 0.20 |
Gm8302 |
predicted gene 8302 |
8135 |
0.2 |
| chrX_142490315_142490613 | 0.20 |
Gm25915 |
predicted gene, 25915 |
5017 |
0.21 |
| chr2_8124945_8125169 | 0.20 |
Gm13254 |
predicted gene 13254 |
22808 |
0.29 |
| chr2_115093304_115093458 | 0.20 |
Gm28493 |
predicted gene 28493 |
42169 |
0.17 |
| chr5_45495684_45495835 | 0.20 |
Lap3 |
leucine aminopeptidase 3 |
2233 |
0.21 |
| chr16_52701782_52701933 | 0.20 |
4930404A05Rik |
RIKEN cDNA 4930404A05 gene |
113887 |
0.06 |
| chr10_24227101_24227252 | 0.20 |
Moxd1 |
monooxygenase, DBH-like 1 |
3631 |
0.24 |
| chr6_29474717_29474868 | 0.20 |
Atp6v1fnb |
Atp6v1f neighbor |
3355 |
0.13 |
| chr10_52273135_52273286 | 0.20 |
Dcbld1 |
discoidin, CUB and LCCL domain containing 1 |
39573 |
0.11 |
| chr1_86633273_86633493 | 0.20 |
Gm22955 |
predicted gene, 22955 |
4225 |
0.14 |
| chr14_70331481_70331660 | 0.20 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
52 |
0.96 |
| chr17_86150939_86151254 | 0.19 |
Srbd1 |
S1 RNA binding domain 1 |
5921 |
0.21 |
| chr7_105592397_105592767 | 0.19 |
Hpx |
hemopexin |
7530 |
0.09 |
| chr1_93030160_93030324 | 0.19 |
Kif1a |
kinesin family member 1A |
3037 |
0.18 |
| chr15_35268847_35269014 | 0.19 |
Osr2 |
odd-skipped related 2 |
27168 |
0.17 |
| chr10_56598323_56598474 | 0.19 |
Gm9795 |
predicted pseudogene 9795 |
57796 |
0.14 |
| chr7_92640281_92640449 | 0.19 |
6430511E19Rik |
RIKEN cDNA 6430511E19 gene |
416 |
0.55 |
| chr9_15653065_15653239 | 0.19 |
Gm7642 |
predicted gene 7642 |
8869 |
0.15 |
| chr10_41893722_41893893 | 0.19 |
Sesn1 |
sestrin 1 |
4566 |
0.25 |
| chr11_69663925_69664238 | 0.19 |
Mir1934 |
microRNA 1934 |
1038 |
0.16 |
| chr11_121085268_121085439 | 0.19 |
Sectm1a |
secreted and transmembrane 1A |
4133 |
0.1 |
| chr19_7456855_7457030 | 0.19 |
Rtn3 |
reticulon 3 |
26260 |
0.1 |
| chr8_9501766_9501947 | 0.19 |
4930435N07Rik |
RIKEN cDNA 4930435N07 gene |
42764 |
0.13 |
| chr14_9608900_9609103 | 0.19 |
Gm48371 |
predicted gene, 48371 |
139446 |
0.05 |
| chr4_62629625_62629794 | 0.19 |
Gm11209 |
predicted gene 11209 |
4003 |
0.2 |
| chr5_143056564_143056750 | 0.19 |
Gm43378 |
predicted gene 43378 |
6614 |
0.15 |
| chr9_118101918_118102083 | 0.19 |
Azi2 |
5-azacytidine induced gene 2 |
42944 |
0.13 |
| chr3_72659248_72659399 | 0.19 |
Gm37901 |
predicted gene, 37901 |
54975 |
0.16 |
| chr3_129617903_129618076 | 0.19 |
Egf |
epidermal growth factor |
63557 |
0.08 |
| chr19_10690326_10690666 | 0.19 |
Vps37c |
vacuolar protein sorting 37C |
1731 |
0.21 |
| chr1_55529058_55529209 | 0.19 |
Gm37382 |
predicted gene, 37382 |
30327 |
0.18 |
| chr13_64349576_64349754 | 0.18 |
Gm47123 |
predicted gene, 47123 |
11387 |
0.1 |
| chr5_54060085_54060253 | 0.18 |
Stim2 |
stromal interaction molecule 2 |
14568 |
0.23 |
| chr3_116189519_116189691 | 0.18 |
Gm31651 |
predicted gene, 31651 |
10803 |
0.15 |
| chr18_12466149_12466310 | 0.18 |
Gm29199 |
predicted gene 29199 |
33233 |
0.13 |
| chr16_30525931_30526082 | 0.18 |
Tmem44 |
transmembrane protein 44 |
14903 |
0.19 |
| chr3_57504904_57505080 | 0.18 |
Gm16016 |
predicted gene 16016 |
23240 |
0.17 |
| chr12_105019429_105019596 | 0.18 |
Gm47648 |
predicted gene, 47648 |
1847 |
0.2 |
| chr3_135931021_135931195 | 0.18 |
Gm31243 |
predicted gene, 31243 |
2876 |
0.22 |
| chr13_107600592_107600794 | 0.18 |
Gm32090 |
predicted gene, 32090 |
12156 |
0.22 |
| chr6_91488990_91489149 | 0.18 |
Tmem43 |
transmembrane protein 43 |
12469 |
0.1 |
| chr1_100496636_100496787 | 0.18 |
Gm18700 |
predicted gene, 18700 |
100364 |
0.08 |
| chr11_16830427_16830830 | 0.18 |
Egfros |
epidermal growth factor receptor, opposite strand |
74 |
0.98 |
| chr1_172903470_172903651 | 0.18 |
Apcs |
serum amyloid P-component |
8519 |
0.16 |
| chr7_16639158_16639452 | 0.18 |
Gm29725 |
predicted gene, 29725 |
8484 |
0.09 |
| chr6_35273676_35273858 | 0.18 |
Slc13a4 |
solute carrier family 13 (sodium/sulfate symporters), member 4 |
14407 |
0.14 |
| chr5_90424369_90424630 | 0.18 |
Gm43363 |
predicted gene 43363 |
27226 |
0.13 |
| chr16_22358863_22359092 | 0.18 |
Etv5 |
ets variant 5 |
42852 |
0.11 |
| chr14_63899614_63899769 | 0.18 |
Pinx1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
78 |
0.98 |
| chr8_104110853_104111054 | 0.18 |
Cdh5 |
cadherin 5 |
354 |
0.82 |
| chr16_43335246_43335402 | 0.18 |
Gm15711 |
predicted gene 15711 |
22730 |
0.14 |
| chr1_115990300_115990467 | 0.18 |
Cntnap5a |
contactin associated protein-like 5A |
305627 |
0.01 |
| chr11_16819865_16820197 | 0.18 |
Egfros |
epidermal growth factor receptor, opposite strand |
10671 |
0.22 |
| chr4_102513252_102513556 | 0.17 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
43007 |
0.2 |
| chr12_102748209_102748378 | 0.17 |
Tmem251 |
transmembrane protein 251 |
4531 |
0.09 |
| chr11_103262520_103262746 | 0.17 |
Map3k14 |
mitogen-activated protein kinase kinase kinase 14 |
4839 |
0.14 |
| chr1_35759768_35759937 | 0.17 |
Gm25634 |
predicted gene, 25634 |
1750 |
0.42 |
| chr15_4561546_4561984 | 0.17 |
C6 |
complement component 6 |
165410 |
0.03 |
| chr2_21245334_21245495 | 0.17 |
Gm13378 |
predicted gene 13378 |
1309 |
0.45 |
| chr11_118265609_118265912 | 0.17 |
Usp36 |
ubiquitin specific peptidase 36 |
288 |
0.89 |
| chr4_101742085_101742265 | 0.17 |
Lepr |
leptin receptor |
24768 |
0.19 |
| chr7_46755251_46755407 | 0.17 |
Saa2 |
serum amyloid A 2 |
3458 |
0.11 |
| chr15_3743269_3743441 | 0.17 |
Gm4823 |
predicted gene 4823 |
3520 |
0.34 |
| chr16_45365461_45365847 | 0.17 |
Cd200 |
CD200 antigen |
34658 |
0.14 |
| chr13_51833684_51833946 | 0.17 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
12863 |
0.21 |
| chr2_59670423_59670592 | 0.17 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
23696 |
0.24 |
| chr19_37213872_37214035 | 0.17 |
Marchf5 |
membrane associated ring-CH-type finger 5 |
3237 |
0.17 |
| chr17_15701795_15701946 | 0.17 |
Chd1 |
chromodomain helicase DNA binding protein 1 |
3097 |
0.26 |
| chr12_83927956_83928132 | 0.17 |
Numb |
NUMB endocytic adaptor protein |
6110 |
0.12 |
| chr4_135989135_135989286 | 0.17 |
Pithd1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
1946 |
0.16 |
| chr5_53196740_53196918 | 0.17 |
Sel1l3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
16486 |
0.19 |
| chr5_128579419_128579592 | 0.17 |
Fzd10os |
frizzled class receptor 10, opposite strand |
5873 |
0.17 |
| chr18_64079634_64079810 | 0.17 |
Gm6974 |
predicted gene 6974 |
1698 |
0.42 |
| chr18_6501445_6501680 | 0.17 |
Epc1 |
enhancer of polycomb homolog 1 |
10706 |
0.16 |
| chr8_126591993_126592171 | 0.17 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
1904 |
0.39 |
| chr2_120170818_120170979 | 0.17 |
Ehd4 |
EH-domain containing 4 |
16292 |
0.15 |
| chr15_75007426_75007737 | 0.17 |
Gm28068 |
predicted gene 28068 |
278 |
0.8 |
| chr2_6333653_6333804 | 0.17 |
AL845275.1 |
novel protein |
10648 |
0.19 |
| chr13_54988316_54988535 | 0.17 |
Unc5a |
unc-5 netrin receptor A |
8114 |
0.16 |
| chr12_104175756_104175908 | 0.17 |
Serpina3d-ps |
serine (or cysteine) peptidase inhibitor, clade A, member 3D, pseudogene |
2464 |
0.15 |
| chr8_86807888_86808046 | 0.17 |
N4bp1 |
NEDD4 binding protein 1 |
36078 |
0.13 |
| chr7_71289589_71289818 | 0.17 |
Gm29328 |
predicted gene 29328 |
25428 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.0 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.0 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.0 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME G1 S TRANSITION | Genes involved in G1/S Transition |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |