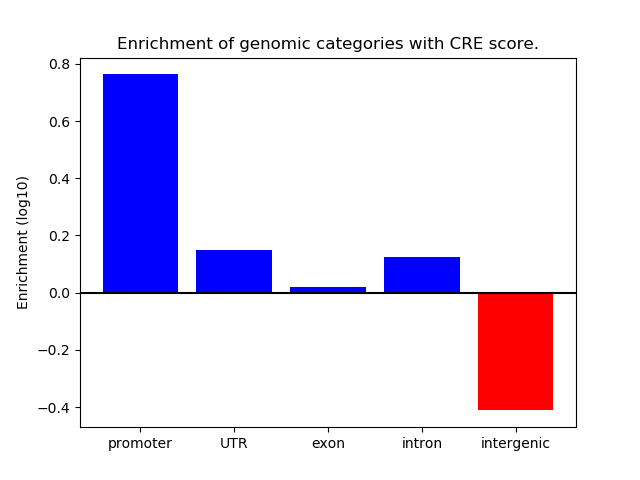
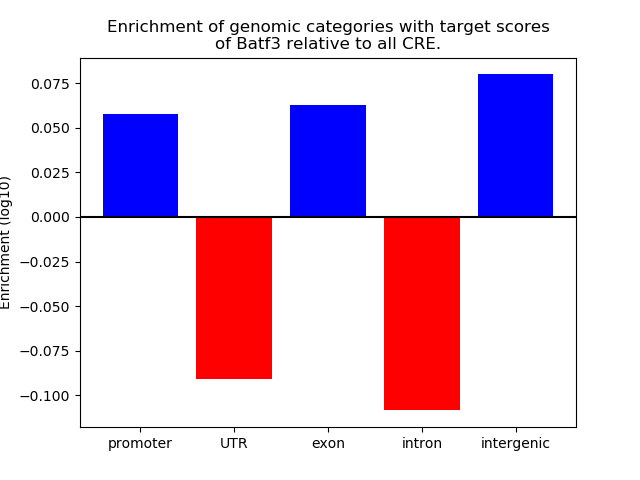
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
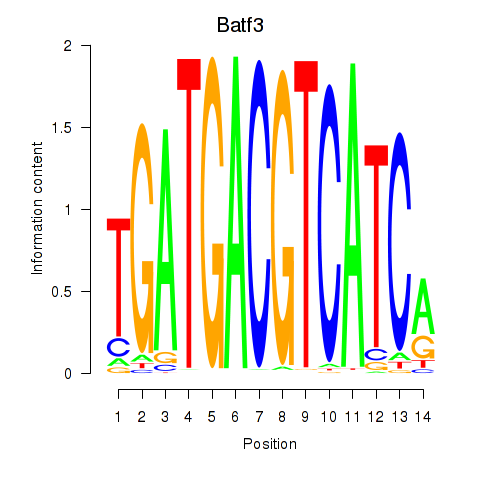
Results for Batf3
Z-value: 0.69
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.4 | basic leucine zipper transcription factor, ATF-like 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_191092186_191092358 | Batf3 | 5575 | 0.118448 | 0.60 | 2.0e-01 | Click! |
Activity of the Batf3 motif across conditions
Conditions sorted by the z-value of the Batf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
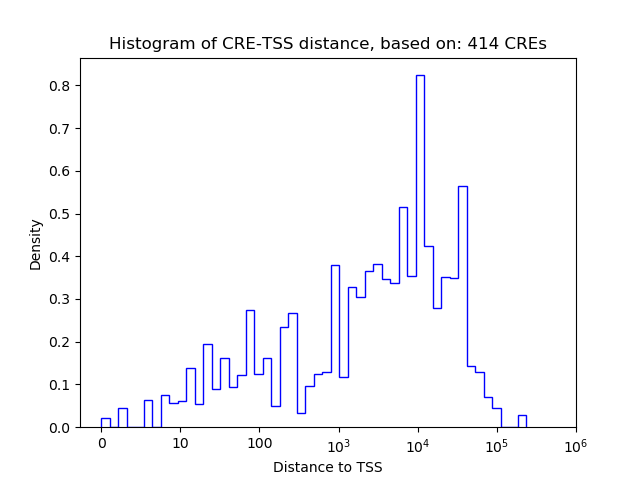
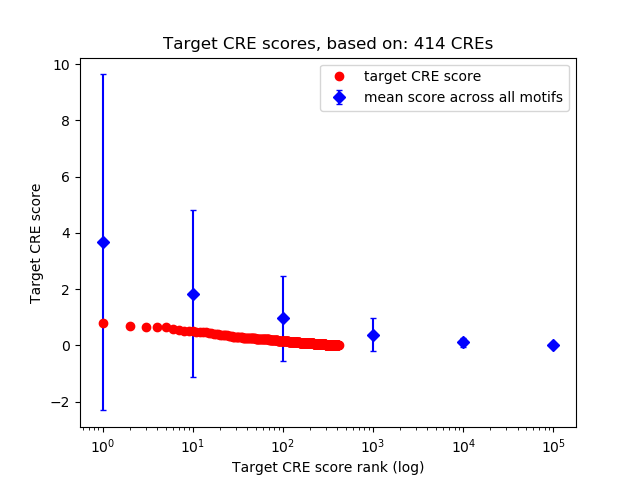
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_19933617_19933791 | 0.80 |
Igsf23 |
immunoglobulin superfamily, member 23 |
11194 |
0.07 |
| chr11_7201515_7201726 | 0.70 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
3838 |
0.2 |
| chr13_113168399_113168569 | 0.66 |
Gzmk |
granzyme K |
12413 |
0.12 |
| chr11_109733399_109733703 | 0.66 |
Fam20a |
family with sequence similarity 20, member A |
11272 |
0.17 |
| chr10_94882831_94882985 | 0.65 |
Gm48718 |
predicted gene, 48718 |
884 |
0.59 |
| chr9_43708878_43709035 | 0.58 |
Gm30015 |
predicted gene, 30015 |
12041 |
0.15 |
| chr12_82225847_82226035 | 0.55 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
30644 |
0.19 |
| chr7_71872986_71873347 | 0.52 |
4930441H08Rik |
RIKEN cDNA 4930441H08 gene |
36527 |
0.18 |
| chr15_91103922_91104096 | 0.51 |
Gm7167 |
predicted gene 7167 |
25888 |
0.16 |
| chr15_66911047_66911248 | 0.50 |
Ccn4 |
cellular communication network factor 4 |
1836 |
0.32 |
| chr4_150512381_150512639 | 0.49 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
24117 |
0.2 |
| chr6_54117349_54117506 | 0.48 |
Chn2 |
chimerin 2 |
19707 |
0.18 |
| chr16_28881932_28882083 | 0.47 |
Mb21d2 |
Mab-21 domain containing 2 |
45640 |
0.17 |
| chr12_109988471_109988667 | 0.46 |
Gm34667 |
predicted gene, 34667 |
35304 |
0.09 |
| chrX_162902296_162902497 | 0.45 |
Ctps2 |
cytidine 5'-triphosphate synthase 2 |
225 |
0.89 |
| chr9_124424931_124425085 | 0.43 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
854 |
0.55 |
| chr3_130696475_130696626 | 0.42 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
72 |
0.96 |
| chr6_120807534_120807700 | 0.41 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
5195 |
0.15 |
| chr5_142960238_142960403 | 0.39 |
Fscn1 |
fascin actin-bundling protein 1 |
23 |
0.97 |
| chr2_4919371_4919703 | 0.37 |
Phyh |
phytanoyl-CoA hydroxylase |
457 |
0.76 |
| chr10_108333685_108334010 | 0.37 |
Pawr |
PRKC, apoptosis, WT1, regulator |
1030 |
0.57 |
| chr1_180422529_180422728 | 0.37 |
Stum |
mechanosensory transduction mediator |
39984 |
0.1 |
| chr2_32167931_32168112 | 0.37 |
Gm27805 |
predicted gene, 27805 |
5150 |
0.13 |
| chr17_35251426_35251588 | 0.35 |
Mir8094 |
microRNA 8094 |
218 |
0.76 |
| chr6_86669367_86669537 | 0.35 |
Mxd1 |
MAX dimerization protein 1 |
291 |
0.82 |
| chr2_125470672_125470852 | 0.33 |
Gm9913 |
predicted gene 9913 |
34327 |
0.17 |
| chr15_69041867_69042024 | 0.32 |
Gm49422 |
predicted gene, 49422 |
82414 |
0.1 |
| chr9_79213145_79213400 | 0.31 |
Gm47499 |
predicted gene, 47499 |
11265 |
0.17 |
| chr5_65955908_65956072 | 0.31 |
4930480C01Rik |
RIKEN cDNA 4930480C01 gene |
4685 |
0.12 |
| chr6_86472269_86472431 | 0.31 |
C87436 |
expressed sequence C87436 |
8310 |
0.08 |
| chr18_12786708_12786879 | 0.29 |
Osbpl1a |
oxysterol binding protein-like 1A |
19656 |
0.15 |
| chr5_122636983_122637314 | 0.29 |
P2rx7 |
purinergic receptor P2X, ligand-gated ion channel, 7 |
6763 |
0.14 |
| chr5_32195986_32196216 | 0.28 |
Gm9555 |
predicted gene 9555 |
10604 |
0.15 |
| chr2_71327268_71327425 | 0.28 |
Slc25a12 |
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
1494 |
0.38 |
| chr17_84119689_84119859 | 0.28 |
Gm19696 |
predicted gene, 19696 |
16186 |
0.15 |
| chr1_155050256_155050434 | 0.27 |
Gm29282 |
predicted gene 29282 |
6834 |
0.17 |
| chr5_123854435_123854768 | 0.27 |
Hcar2 |
hydroxycarboxylic acid receptor 2 |
10898 |
0.13 |
| chr17_26538205_26538356 | 0.27 |
Gm50275 |
predicted gene, 50275 |
53 |
0.95 |
| chr16_13354001_13354171 | 0.27 |
Mrtfb |
myocardin related transcription factor B |
4335 |
0.25 |
| chr12_71045001_71045164 | 0.27 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
3259 |
0.22 |
| chr12_75307960_75308125 | 0.26 |
Rhoj |
ras homolog family member J |
280 |
0.95 |
| chr2_29828898_29829065 | 0.26 |
Urm1 |
ubiquitin related modifier 1 |
1586 |
0.22 |
| chr15_78450182_78450345 | 0.26 |
Tmprss6 |
transmembrane serine protease 6 |
5884 |
0.11 |
| chr3_127553258_127553423 | 0.26 |
Larp7 |
La ribonucleoprotein domain family, member 7 |
7 |
0.64 |
| chr15_62169758_62169909 | 0.26 |
Pvt1 |
Pvt1 oncogene |
8342 |
0.27 |
| chr15_76579147_76579305 | 0.26 |
Gm49413 |
predicted gene, 49413 |
1401 |
0.18 |
| chr10_3573083_3573240 | 0.25 |
Iyd |
iodotyrosine deiodinase |
32873 |
0.22 |
| chr15_66932473_66932630 | 0.25 |
Ndrg1 |
N-myc downstream regulated gene 1 |
1918 |
0.28 |
| chr11_19924205_19924473 | 0.25 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
36 |
0.99 |
| chrX_13070891_13071062 | 0.24 |
Usp9x |
ubiquitin specific peptidase 9, X chromosome |
522 |
0.79 |
| chr11_60133979_60134158 | 0.24 |
Rai1 |
retinoic acid induced 1 |
6014 |
0.16 |
| chr10_63122682_63122839 | 0.23 |
Mypn |
myopalladin |
76 |
0.96 |
| chr5_134555490_134555833 | 0.23 |
Gm42884 |
predicted gene 42884 |
876 |
0.4 |
| chr14_26217468_26217634 | 0.23 |
Gm19015 |
predicted gene, 19015 |
2359 |
0.19 |
| chr11_98785850_98786019 | 0.23 |
Msl1 |
male specific lethal 1 |
9582 |
0.09 |
| chr14_25938104_25938268 | 0.23 |
Cphx1 |
cytoplasmic polyadenylated homeobox 1 |
118 |
0.94 |
| chr14_26077857_26078021 | 0.23 |
Gm19014 |
predicted gene, 19014 |
2357 |
0.16 |
| chr9_102718313_102718471 | 0.22 |
Amotl2 |
angiomotin-like 2 |
37 |
0.96 |
| chr4_141301124_141301279 | 0.22 |
Epha2 |
Eph receptor A2 |
39 |
0.96 |
| chr17_33760685_33760872 | 0.22 |
Rab11b |
RAB11B, member RAS oncogene family |
248 |
0.75 |
| chr6_120932679_120932852 | 0.22 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
2770 |
0.17 |
| chr11_94044111_94044443 | 0.22 |
Spag9 |
sperm associated antigen 9 |
9 |
0.98 |
| chr13_56461581_56461925 | 0.22 |
Il9 |
interleukin 9 |
20493 |
0.14 |
| chr13_51871249_51871406 | 0.22 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
24583 |
0.2 |
| chr12_84279002_84279173 | 0.21 |
Ptgr2 |
prostaglandin reductase 2 |
6145 |
0.12 |
| chr10_19948695_19948879 | 0.21 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
2268 |
0.33 |
| chr1_75168548_75168709 | 0.21 |
Zfand2b |
zinc finger, AN1 type domain 2B |
18 |
0.93 |
| chr4_133928748_133928902 | 0.21 |
Hmgn2 |
high mobility group nucleosomal binding domain 2 |
38418 |
0.08 |
| chr9_103234188_103234350 | 0.21 |
Trf |
transferrin |
3825 |
0.2 |
| chr13_22043005_22043165 | 0.21 |
H2bc11 |
H2B clustered histone 11 |
129 |
0.45 |
| chr14_25346096_25346426 | 0.21 |
Gm26660 |
predicted gene, 26660 |
32374 |
0.17 |
| chr16_52034396_52034559 | 0.20 |
Cblb |
Casitas B-lineage lymphoma b |
2260 |
0.37 |
| chr16_10976237_10976411 | 0.20 |
Litaf |
LPS-induced TN factor |
745 |
0.51 |
| chr9_43545212_43545661 | 0.20 |
Gm36855 |
predicted gene, 36855 |
24432 |
0.17 |
| chr17_28013343_28013511 | 0.20 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
6082 |
0.13 |
| chr6_124811055_124811230 | 0.19 |
Tpi1 |
triosephosphate isomerase 1 |
744 |
0.35 |
| chr5_139484483_139484643 | 0.19 |
Zfand2a |
zinc finger, AN1-type domain 2A |
14 |
0.97 |
| chr16_38940739_38940891 | 0.19 |
Gm22500 |
predicted gene, 22500 |
13606 |
0.21 |
| chr8_108743266_108743566 | 0.18 |
Gm38042 |
predicted gene, 38042 |
5823 |
0.26 |
| chr18_74120787_74120938 | 0.18 |
Mapk4 |
mitogen-activated protein kinase 4 |
55503 |
0.1 |
| chr15_36960415_36960763 | 0.18 |
Gm34590 |
predicted gene, 34590 |
21725 |
0.14 |
| chr5_73168897_73169054 | 0.18 |
Fryl |
FRY like transcription coactivator |
535 |
0.7 |
| chr9_55333127_55333288 | 0.18 |
Tmem266 |
transmembrane protein 266 |
6 |
0.98 |
| chr17_56476460_56476653 | 0.18 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
73 |
0.96 |
| chr7_45526245_45526410 | 0.18 |
Plekha4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
3 |
0.51 |
| chr8_88633569_88633732 | 0.17 |
Snx20 |
sorting nexin 20 |
2451 |
0.26 |
| chr9_118674269_118674608 | 0.17 |
Itga9 |
integrin alpha 9 |
11397 |
0.23 |
| chr15_3268359_3268542 | 0.17 |
Selenop |
selenoprotein P |
97 |
0.97 |
| chr7_138846212_138846376 | 0.17 |
Mapk1ip1 |
mitogen-activated protein kinase 1 interacting protein 1 |
21 |
0.53 |
| chr2_103846113_103846314 | 0.17 |
Gm13879 |
predicted gene 13879 |
2557 |
0.13 |
| chr11_115462203_115462381 | 0.17 |
Slc16a5 |
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
182 |
0.83 |
| chr1_184852008_184852353 | 0.16 |
Mtarc2 |
mitochondrial amidoxime reducing component 2 |
5729 |
0.17 |
| chr10_82354496_82354666 | 0.16 |
Gm4924 |
predicted gene 4924 |
265 |
0.91 |
| chr3_13783011_13783180 | 0.16 |
Ralyl |
RALY RNA binding protein-like |
6247 |
0.24 |
| chr12_54459633_54459834 | 0.16 |
Gm7557 |
predicted gene 7557 |
29335 |
0.13 |
| chr17_29599607_29599789 | 0.16 |
Tbc1d22bos |
TBC1 domain family, member 22B, opposite strand |
3393 |
0.11 |
| chr6_128799347_128799643 | 0.16 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
10280 |
0.1 |
| chr3_129536318_129536484 | 0.15 |
Gm43072 |
predicted gene 43072 |
1740 |
0.29 |
| chr4_155235347_155235531 | 0.15 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
12847 |
0.15 |
| chr8_69192982_69193338 | 0.15 |
Lzts1 |
leucine zipper, putative tumor suppressor 1 |
8935 |
0.15 |
| chr19_43898660_43898822 | 0.15 |
Dnmbp |
dynamin binding protein |
8050 |
0.14 |
| chr14_77034577_77034753 | 0.15 |
Lacc1 |
laccase domain containing 1 |
1416 |
0.39 |
| chr5_130081319_130081518 | 0.15 |
Tpst1 |
protein-tyrosine sulfotransferase 1 |
2048 |
0.24 |
| chr10_75121130_75121291 | 0.15 |
Bcr |
BCR activator of RhoGEF and GTPase |
39150 |
0.15 |
| chr16_30732879_30733030 | 0.15 |
Gm49754 |
predicted gene, 49754 |
60301 |
0.1 |
| chr2_58794623_58794807 | 0.15 |
Upp2 |
uridine phosphorylase 2 |
29390 |
0.17 |
| chr15_66996996_66997162 | 0.15 |
Ndrg1 |
N-myc downstream regulated gene 1 |
27439 |
0.15 |
| chr14_68048482_68048633 | 0.15 |
Gm31107 |
predicted gene, 31107 |
2773 |
0.28 |
| chr17_73881220_73881413 | 0.14 |
Xdh |
xanthine dehydrogenase |
5434 |
0.2 |
| chr2_133550506_133550686 | 0.14 |
Bmp2 |
bone morphogenetic protein 2 |
1563 |
0.4 |
| chr13_23530925_23531217 | 0.14 |
H4c8 |
H4 clustered histone 8 |
21 |
0.59 |
| chr5_123980690_123981011 | 0.14 |
Hip1r |
huntingtin interacting protein 1 related |
596 |
0.59 |
| chr4_40703144_40703305 | 0.14 |
Aptx |
aprataxin |
30 |
0.96 |
| chr1_155063987_155064316 | 0.14 |
Gm29282 |
predicted gene 29282 |
6972 |
0.17 |
| chr9_106170725_106171009 | 0.14 |
Wdr82 |
WD repeat domain containing 82 |
61 |
0.94 |
| chr11_112821671_112821851 | 0.13 |
4933434M16Rik |
RIKEN cDNA 4933434M16 gene |
3393 |
0.25 |
| chr15_81190841_81191005 | 0.13 |
4930483J18Rik |
RIKEN cDNA 4930483J18 gene |
70 |
0.63 |
| chr2_125123073_125123271 | 0.13 |
Myef2 |
myelin basic protein expression factor 2, repressor |
242 |
0.89 |
| chr8_127440670_127440852 | 0.13 |
Pard3 |
par-3 family cell polarity regulator |
6985 |
0.31 |
| chr5_24383598_24383768 | 0.13 |
Gm15587 |
predicted gene 15587 |
69 |
0.93 |
| chr11_121086049_121086611 | 0.13 |
Sectm1a |
secreted and transmembrane 1A |
5110 |
0.09 |
| chr6_115465780_115466186 | 0.13 |
Gm44079 |
predicted gene, 44079 |
993 |
0.54 |
| chrX_104293728_104293906 | 0.12 |
Vcp-rs |
valosin containing protein, related sequence |
54417 |
0.12 |
| chr13_19622939_19623114 | 0.12 |
Sfrp4 |
secreted frizzled-related protein 4 |
78 |
0.97 |
| chr8_3414105_3414577 | 0.12 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
12359 |
0.15 |
| chrX_168941822_168941984 | 0.12 |
Arhgap6 |
Rho GTPase activating protein 6 |
94708 |
0.08 |
| chr17_34119461_34119627 | 0.12 |
H2-DMa |
histocompatibility 2, class II, locus DMa |
3 |
0.87 |
| chr14_20226275_20226463 | 0.12 |
Gm48395 |
predicted gene, 48395 |
10867 |
0.12 |
| chr6_17532117_17532312 | 0.11 |
Met |
met proto-oncogene |
2787 |
0.33 |
| chr13_55425993_55426169 | 0.11 |
F12 |
coagulation factor XII (Hageman factor) |
698 |
0.47 |
| chr13_67306400_67306560 | 0.11 |
Zfp457 |
zinc finger protein 457 |
5 |
0.94 |
| chr6_134160918_134161473 | 0.11 |
Gm17088 |
predicted gene 17088 |
10812 |
0.2 |
| chr5_123436059_123436381 | 0.11 |
Mlxip |
MLX interacting protein |
9819 |
0.09 |
| chr17_65701951_65702157 | 0.11 |
Gm49867 |
predicted gene, 49867 |
40476 |
0.12 |
| chr19_61226557_61226761 | 0.11 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
24 |
0.96 |
| chrY_2644529_2644731 | 0.11 |
Uba1y-ps1 |
ubiquitin-activating enzyme, Chr Y, pseudogene 1 |
2670 |
0.2 |
| chr17_72399518_72399681 | 0.11 |
Gm24736 |
predicted gene, 24736 |
99819 |
0.07 |
| chrX_106190211_106190376 | 0.11 |
Pgk1 |
phosphoglycerate kinase 1 |
1335 |
0.35 |
| chr5_100583214_100583366 | 0.10 |
Gm43510 |
predicted gene 43510 |
9466 |
0.14 |
| chr3_21935997_21936168 | 0.10 |
Gm43674 |
predicted gene 43674 |
62386 |
0.12 |
| chr11_102742573_102742724 | 0.10 |
Gm16342 |
predicted gene 16342 |
10243 |
0.11 |
| chr15_7137772_7137928 | 0.10 |
Lifr |
LIF receptor alpha |
2692 |
0.38 |
| chrY_2681513_2681716 | 0.10 |
Gm18798 |
predicted gene, 18798 |
2670 |
0.2 |
| chr8_70213088_70213257 | 0.10 |
Slc25a42 |
solute carrier family 25, member 42 |
867 |
0.42 |
| chr9_107612764_107612937 | 0.10 |
Gnai2 |
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
3197 |
0.08 |
| chr1_86606018_86606363 | 0.10 |
Cops7b |
COP9 signalosome subunit 7B |
9450 |
0.12 |
| chr2_109280667_109280830 | 0.10 |
Mettl15 |
methyltransferase like 15 |
0 |
0.5 |
| chrX_101953814_101953986 | 0.10 |
Nhsl2 |
NHS-like 2 |
7897 |
0.18 |
| chr11_116657951_116658105 | 0.10 |
Prcd |
photoreceptor disc component |
872 |
0.37 |
| chr19_29037790_29038010 | 0.10 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
8345 |
0.11 |
| chr1_131079599_131079858 | 0.10 |
Gm25549 |
predicted gene, 25549 |
5977 |
0.14 |
| chr5_36691549_36691700 | 0.10 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
4313 |
0.16 |
| chr16_34939309_34939471 | 0.10 |
1700119H24Rik |
RIKEN cDNA 1700119H24 gene |
61 |
0.97 |
| chr2_158316475_158316655 | 0.10 |
Lbp |
lipopolysaccharide binding protein |
2359 |
0.18 |
| chr2_38111461_38111697 | 0.10 |
Gm44291 |
predicted gene, 44291 |
4283 |
0.25 |
| chr7_16119633_16119956 | 0.10 |
Kptn |
kaptin |
101 |
0.95 |
| chr7_100591363_100591708 | 0.10 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
109 |
0.93 |
| chr7_44672289_44672471 | 0.10 |
Myh14 |
myosin, heavy polypeptide 14 |
1537 |
0.22 |
| chr15_86189223_86189384 | 0.10 |
Cerk |
ceramide kinase |
3162 |
0.2 |
| chr17_34586879_34587070 | 0.10 |
Notch4 |
notch 4 |
29 |
0.91 |
| chr19_38365309_38365465 | 0.10 |
Gm50155 |
predicted gene, 50155 |
20127 |
0.13 |
| chr12_79561056_79561219 | 0.09 |
Rad51b |
RAD51 paralog B |
233784 |
0.02 |
| chr16_93710965_93711118 | 0.09 |
Dop1b |
DOP1 leucine zipper like protein B |
863 |
0.55 |
| chr4_142017836_142017987 | 0.09 |
4930455G09Rik |
RIKEN cDNA 4930455G09 gene |
13 |
0.96 |
| chr11_20200974_20201192 | 0.09 |
Rab1a |
RAB1A, member RAS oncogene family |
349 |
0.85 |
| chr6_128764252_128764459 | 0.09 |
Gm43907 |
predicted gene, 43907 |
19711 |
0.09 |
| chr1_161035922_161036116 | 0.09 |
Gas5 |
growth arrest specific 5 |
1 |
0.38 |
| chr4_107303606_107303802 | 0.09 |
Dio1 |
deiodinase, iodothyronine, type I |
3362 |
0.15 |
| chr11_69417573_69417747 | 0.09 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
3985 |
0.1 |
| chr8_69716419_69716582 | 0.09 |
Zfp869 |
zinc finger protein 869 |
14 |
0.97 |
| chr9_121348150_121348320 | 0.09 |
Gm47092 |
predicted gene, 47092 |
1679 |
0.32 |
| chr18_38962798_38962983 | 0.09 |
Gm5820 |
predicted gene 5820 |
7181 |
0.19 |
| chr13_34077943_34078110 | 0.09 |
Tubb2a |
tubulin, beta 2A class IIA |
19 |
0.96 |
| chr2_127444501_127444657 | 0.09 |
Fahd2a |
fumarylacetoacetate hydrolase domain containing 2A |
14 |
0.97 |
| chr11_88242033_88242214 | 0.09 |
Gm38534 |
predicted gene, 38534 |
11245 |
0.17 |
| chr7_16296893_16297072 | 0.09 |
Ccdc9 |
coiled-coil domain containing 9 |
10187 |
0.11 |
| chr5_134269920_134270079 | 0.09 |
Gm43481 |
predicted gene 43481 |
737 |
0.54 |
| chr10_18237308_18237472 | 0.09 |
Gm10827 |
predicted gene 10827 |
2024 |
0.26 |
| chr16_55698500_55698846 | 0.09 |
Gm19771 |
predicted gene, 19771 |
41245 |
0.16 |
| chr4_59177698_59177849 | 0.09 |
Ugcg |
UDP-glucose ceramide glucosyltransferase |
11484 |
0.15 |
| chr7_145111866_145112053 | 0.09 |
Gm45181 |
predicted gene 45181 |
51037 |
0.14 |
| chr2_120154260_120154763 | 0.09 |
Ehd4 |
EH-domain containing 4 |
49 |
0.97 |
| chr9_100599823_100599989 | 0.09 |
Stag1 |
stromal antigen 1 |
2108 |
0.23 |
| chr5_123057781_123058133 | 0.09 |
Gm6444 |
predicted gene 6444 |
8285 |
0.09 |
| chr15_99116561_99116759 | 0.09 |
Spats2 |
spermatogenesis associated, serine-rich 2 |
9256 |
0.09 |
| chr4_155895345_155895496 | 0.08 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
3108 |
0.1 |
| chr10_81182793_81182961 | 0.08 |
Dapk3 |
death-associated protein kinase 3 |
128 |
0.84 |
| chr2_75763382_75763582 | 0.08 |
Gm13657 |
predicted gene 13657 |
13706 |
0.14 |
| chr10_108360046_108360218 | 0.08 |
Gm23105 |
predicted gene, 23105 |
3533 |
0.25 |
| chr19_47317600_47317752 | 0.08 |
Sh3pxd2a |
SH3 and PX domains 2A |
2925 |
0.24 |
| chr7_7298727_7298897 | 0.08 |
Clcn4 |
chloride channel, voltage-sensitive 4 |
6 |
0.55 |
| chr10_121510171_121510329 | 0.08 |
Gm35696 |
predicted gene, 35696 |
148 |
0.93 |
| chr9_124424253_124424419 | 0.08 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
182 |
0.93 |
| chr18_3281039_3281202 | 0.08 |
Crem |
cAMP responsive element modulator |
12 |
0.98 |
| chr4_128610093_128610254 | 0.08 |
Zscan20 |
zinc finger and SCAN domains 20 |
75 |
0.97 |
| chr16_57181297_57181460 | 0.07 |
Tbc1d23 |
TBC1 domain family, member 23 |
1174 |
0.41 |
| chr6_114890473_114890633 | 0.07 |
Vgll4 |
vestigial like family member 4 |
15446 |
0.2 |
| chr11_61274046_61274219 | 0.07 |
Aldh3a2 |
aldehyde dehydrogenase family 3, subfamily A2 |
6668 |
0.17 |
| chr3_96094342_96094503 | 0.07 |
Gm43554 |
predicted gene 43554 |
7551 |
0.1 |
| chr3_104677278_104677438 | 0.07 |
Gm29560 |
predicted gene 29560 |
7348 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0071335 | hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.0 | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process(GO:0009147) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |