Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
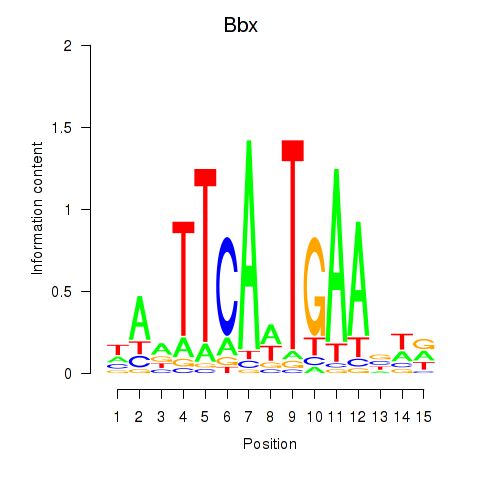
Results for Bbx
Z-value: 1.08
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSMUSG00000022641.9 | bobby sox HMG box containing |
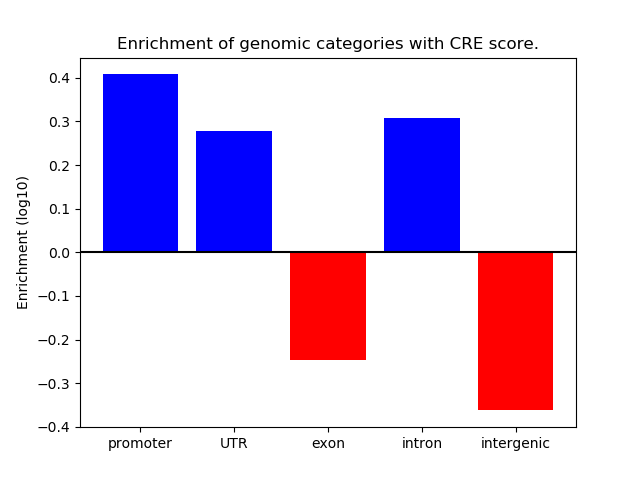
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_50431377_50431947 | Bbx | 647 | 0.811981 | -0.87 | 2.3e-02 | Click! |
| chr16_50429243_50429410 | Bbx | 586 | 0.837552 | -0.86 | 2.6e-02 | Click! |
| chr16_50432097_50432257 | Bbx | 132 | 0.977663 | -0.70 | 1.2e-01 | Click! |
| chr16_50430297_50430475 | Bbx | 474 | 0.881324 | -0.58 | 2.3e-01 | Click! |
| chr16_50364236_50364416 | Bbx | 33333 | 0.229106 | 0.53 | 2.8e-01 | Click! |
Activity of the Bbx motif across conditions
Conditions sorted by the z-value of the Bbx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
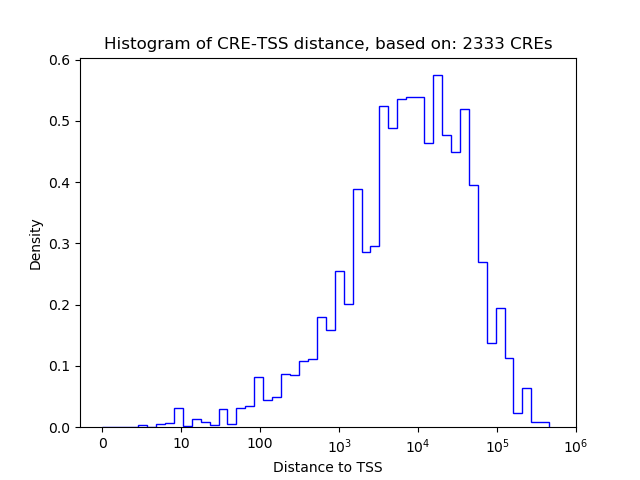
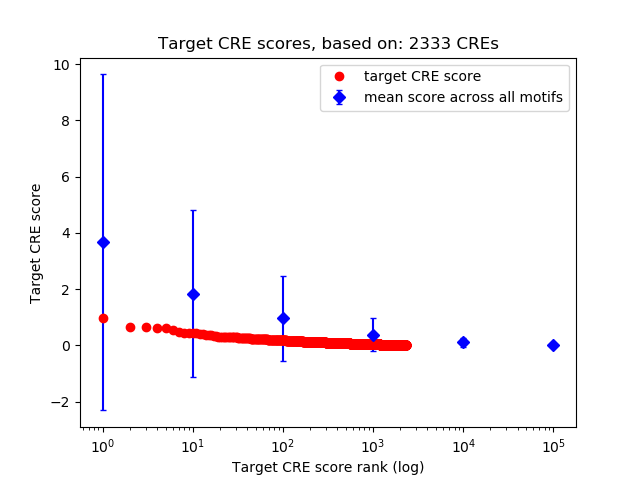
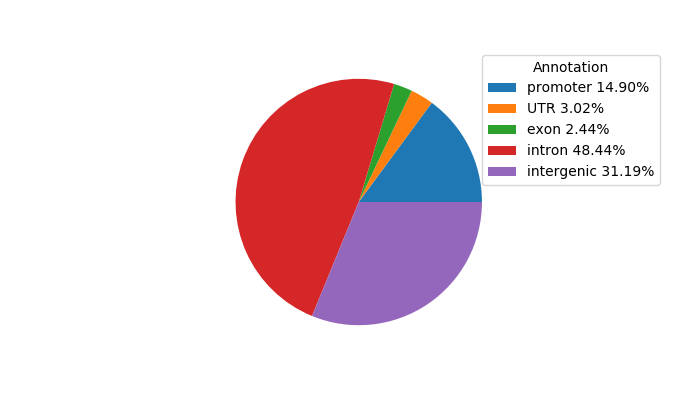
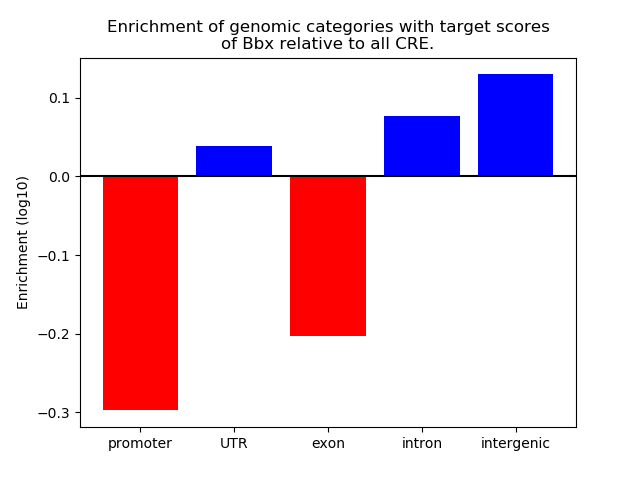
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_105328178_105328374 | 0.97 |
Gm12722 |
predicted gene 12722 |
46670 |
0.19 |
| chr13_4468232_4468415 | 0.65 |
Gm48010 |
predicted gene, 48010 |
1757 |
0.34 |
| chr11_16858925_16859117 | 0.65 |
Egfr |
epidermal growth factor receptor |
19129 |
0.18 |
| chr16_43515591_43515742 | 0.61 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
5358 |
0.27 |
| chr11_16834271_16834692 | 0.61 |
Egfros |
epidermal growth factor receptor, opposite strand |
3779 |
0.26 |
| chr7_87371467_87371654 | 0.55 |
Tyr |
tyrosinase |
121832 |
0.05 |
| chr16_23056284_23056437 | 0.48 |
Kng1 |
kininogen 1 |
1505 |
0.19 |
| chr7_89669589_89669767 | 0.44 |
A230065N10Rik |
RIKEN cDNA A230065N10 gene |
36554 |
0.13 |
| chr11_16860515_16860728 | 0.44 |
Egfr |
epidermal growth factor receptor |
17529 |
0.19 |
| chr8_76152114_76152295 | 0.42 |
Gm45742 |
predicted gene 45742 |
35177 |
0.2 |
| chr6_93395998_93396170 | 0.42 |
Gm44181 |
predicted gene, 44181 |
59473 |
0.12 |
| chr13_62860030_62860181 | 0.41 |
Fbp2 |
fructose bisphosphatase 2 |
1683 |
0.26 |
| chr10_99314465_99314820 | 0.41 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
15476 |
0.14 |
| chr10_99313401_99313555 | 0.38 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
14312 |
0.14 |
| chr9_74891211_74891440 | 0.36 |
Onecut1 |
one cut domain, family member 1 |
24841 |
0.14 |
| chr2_8116897_8117048 | 0.36 |
Gm13254 |
predicted gene 13254 |
30893 |
0.26 |
| chr8_64735601_64735761 | 0.35 |
Msmo1 |
methylsterol monoxygenase 1 |
1889 |
0.29 |
| chr8_44709672_44709838 | 0.35 |
Gm26089 |
predicted gene, 26089 |
107922 |
0.07 |
| chr4_52816035_52816203 | 0.30 |
Olfr275 |
olfactory receptor 275 |
1515 |
0.32 |
| chr3_112820887_112821047 | 0.30 |
Gm43587 |
predicted gene 43587 |
53955 |
0.17 |
| chr10_87549648_87549853 | 0.30 |
Pah |
phenylalanine hydroxylase |
3077 |
0.29 |
| chr16_14950695_14950883 | 0.30 |
Gm33696 |
predicted gene, 33696 |
41094 |
0.14 |
| chr3_98670706_98671055 | 0.29 |
Gm12399 |
predicted gene 12399 |
7451 |
0.14 |
| chr3_27766821_27766972 | 0.29 |
Fndc3b |
fibronectin type III domain containing 3B |
55589 |
0.14 |
| chr1_67153015_67153221 | 0.29 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
30092 |
0.19 |
| chr5_33465811_33466048 | 0.29 |
Gm43851 |
predicted gene 43851 |
28465 |
0.15 |
| chr11_16589151_16589356 | 0.29 |
Gm12663 |
predicted gene 12663 |
46813 |
0.12 |
| chrX_19139471_19139640 | 0.28 |
Mir221 |
microRNA 221 |
6833 |
0.2 |
| chr8_72131379_72131550 | 0.28 |
Tpm4 |
tropomyosin 4 |
1290 |
0.23 |
| chr14_22964972_22965155 | 0.28 |
Gm47601 |
predicted gene, 47601 |
37137 |
0.22 |
| chr13_64205035_64205186 | 0.28 |
Cdc14b |
CDC14 cell division cycle 14B |
4072 |
0.13 |
| chr3_19043040_19043218 | 0.28 |
Gm37979 |
predicted gene, 37979 |
100770 |
0.07 |
| chrX_61144273_61144442 | 0.27 |
Gm24396 |
predicted gene, 24396 |
1005 |
0.5 |
| chr2_73974269_73974420 | 0.27 |
Gm13668 |
predicted gene 13668 |
13675 |
0.22 |
| chr16_32165698_32165859 | 0.26 |
Nrros |
negative regulator of reactive oxygen species |
184 |
0.92 |
| chr11_16877316_16877467 | 0.26 |
Egfr |
epidermal growth factor receptor |
759 |
0.68 |
| chr1_127898839_127898990 | 0.26 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1999 |
0.31 |
| chr10_52234541_52234692 | 0.26 |
Dcbld1 |
discoidin, CUB and LCCL domain containing 1 |
979 |
0.54 |
| chr10_68126588_68126805 | 0.26 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
9930 |
0.25 |
| chr4_54733562_54733749 | 0.25 |
Gm12477 |
predicted gene 12477 |
63463 |
0.11 |
| chr15_81804535_81804686 | 0.25 |
Tef |
thyrotroph embryonic factor |
1784 |
0.19 |
| chr5_150264404_150264568 | 0.25 |
Fry |
FRY microtubule binding protein |
4719 |
0.21 |
| chr8_114776777_114776930 | 0.24 |
Wwox |
WW domain-containing oxidoreductase |
64686 |
0.12 |
| chr1_9578333_9578671 | 0.24 |
Gm6161 |
predicted gene 6161 |
3954 |
0.16 |
| chr3_129526989_129527140 | 0.24 |
Gm35986 |
predicted gene, 35986 |
5045 |
0.17 |
| chr7_68975121_68975306 | 0.23 |
Gm44691 |
predicted gene 44691 |
14191 |
0.21 |
| chr8_45630758_45630927 | 0.23 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
2641 |
0.27 |
| chrX_35957374_35957532 | 0.23 |
Dock11 |
dedicator of cytokinesis 11 |
10056 |
0.25 |
| chr7_4705237_4705398 | 0.23 |
Brsk1 |
BR serine/threonine kinase 1 |
3661 |
0.1 |
| chr13_24546719_24546907 | 0.23 |
Ripor2 |
RHO family interacting cell polarization regulator 2 |
35376 |
0.14 |
| chr5_8159085_8159252 | 0.23 |
Gm21759 |
predicted gene, 21759 |
20468 |
0.16 |
| chr1_164586384_164586561 | 0.23 |
D630023O14Rik |
RIKEN cDNA D630023O14 gene |
11017 |
0.18 |
| chr8_68016078_68016239 | 0.23 |
Gm22018 |
predicted gene, 22018 |
5818 |
0.26 |
| chr19_56600920_56601157 | 0.23 |
Nhlrc2 |
NHL repeat containing 2 |
30241 |
0.15 |
| chr1_102735468_102735657 | 0.23 |
Gm22034 |
predicted gene, 22034 |
217986 |
0.02 |
| chr4_49535104_49535371 | 0.22 |
Aldob |
aldolase B, fructose-bisphosphate |
3629 |
0.16 |
| chr19_8263388_8263539 | 0.22 |
Gm7105 |
predicted gene 7105 |
5876 |
0.17 |
| chr2_42022610_42022815 | 0.22 |
Gm13461 |
predicted gene 13461 |
33883 |
0.23 |
| chr16_23511406_23511565 | 0.22 |
Gm49514 |
predicted gene, 49514 |
2785 |
0.2 |
| chr17_64961928_64962092 | 0.22 |
Gm25344 |
predicted gene, 25344 |
17158 |
0.25 |
| chr19_26823641_26823967 | 0.22 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
103 |
0.97 |
| chr6_146915387_146915718 | 0.21 |
Ppfibp1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
26963 |
0.13 |
| chr16_87671512_87671714 | 0.21 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
27332 |
0.17 |
| chr10_98666150_98666367 | 0.21 |
Gm5427 |
predicted gene 5427 |
33452 |
0.22 |
| chr14_21281587_21281757 | 0.21 |
Adk |
adenosine kinase |
36453 |
0.2 |
| chr16_3877520_3877678 | 0.21 |
Zfp597 |
zinc finger protein 597 |
5203 |
0.09 |
| chr2_84338164_84338328 | 0.21 |
Gm13711 |
predicted gene 13711 |
19935 |
0.19 |
| chr19_7966794_7966947 | 0.21 |
Slc22a27 |
solute carrier family 22, member 27 |
843 |
0.53 |
| chr3_129297386_129297570 | 0.20 |
Enpep |
glutamyl aminopeptidase |
11556 |
0.16 |
| chr12_75694445_75694598 | 0.20 |
Wdr89 |
WD repeat domain 89 |
24984 |
0.18 |
| chr1_61971987_61972299 | 0.20 |
Gm29640 |
predicted gene 29640 |
9064 |
0.27 |
| chr12_103657456_103657664 | 0.20 |
Serpina6 |
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
348 |
0.79 |
| chr13_63345488_63345639 | 0.20 |
Fancc |
Fanconi anemia, complementation group C |
6609 |
0.11 |
| chr2_4933062_4933213 | 0.20 |
Phyh |
phytanoyl-CoA hydroxylase |
4346 |
0.15 |
| chr2_155592205_155592416 | 0.20 |
Gssos1 |
glutathione synthase, opposite strand 1 |
9 |
0.7 |
| chr5_74625167_74625318 | 0.20 |
Lnx1 |
ligand of numb-protein X 1 |
1721 |
0.35 |
| chr17_64740112_64740263 | 0.20 |
Gm17133 |
predicted gene 17133 |
13909 |
0.19 |
| chr7_79311533_79311809 | 0.19 |
Gm39041 |
predicted gene, 39041 |
12646 |
0.14 |
| chr6_37751805_37751968 | 0.19 |
Atp6v0c-ps2 |
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2 |
54200 |
0.13 |
| chr4_76942367_76942518 | 0.19 |
Gm11246 |
predicted gene 11246 |
15131 |
0.21 |
| chr3_61092785_61092961 | 0.19 |
Gm37035 |
predicted gene, 37035 |
89087 |
0.08 |
| chr11_16899488_16899646 | 0.19 |
Egfr |
epidermal growth factor receptor |
5618 |
0.21 |
| chr2_46521470_46521692 | 0.19 |
Gm13470 |
predicted gene 13470 |
78871 |
0.1 |
| chr13_28672871_28673024 | 0.18 |
Mir6368 |
microRNA 6368 |
37926 |
0.2 |
| chr1_67201067_67201218 | 0.18 |
Gm15668 |
predicted gene 15668 |
48058 |
0.14 |
| chr3_150365886_150366049 | 0.18 |
Gm6439 |
predicted gene 6439 |
150114 |
0.05 |
| chr7_72181786_72181957 | 0.18 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
19497 |
0.2 |
| chr3_108902138_108902289 | 0.18 |
Prpf38b |
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
3372 |
0.17 |
| chr6_133973359_133973527 | 0.18 |
Gm8956 |
predicted gene 8956 |
41034 |
0.14 |
| chr1_91442432_91442601 | 0.18 |
Per2 |
period circadian clock 2 |
7349 |
0.12 |
| chr1_67014326_67014505 | 0.18 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
6650 |
0.2 |
| chr2_147949593_147949773 | 0.18 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
26502 |
0.2 |
| chr12_104087488_104087669 | 0.18 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
6929 |
0.1 |
| chr18_47314660_47314858 | 0.18 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
18567 |
0.22 |
| chr11_86705520_86705677 | 0.18 |
Cltc |
clathrin, heavy polypeptide (Hc) |
1901 |
0.3 |
| chr1_52434736_52434915 | 0.18 |
Gm23460 |
predicted gene, 23460 |
9719 |
0.18 |
| chr11_104549691_104549898 | 0.18 |
Cdc27 |
cell division cycle 27 |
598 |
0.47 |
| chr15_7159645_7159840 | 0.18 |
Lifr |
LIF receptor alpha |
5389 |
0.3 |
| chr13_4577869_4578048 | 0.18 |
Akr1c21 |
aldo-keto reductase family 1, member C21 |
801 |
0.61 |
| chr6_28758962_28759125 | 0.17 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
5647 |
0.24 |
| chr5_63938523_63938715 | 0.17 |
Rell1 |
RELT-like 1 |
7705 |
0.17 |
| chr6_17172773_17172927 | 0.17 |
Gm4876 |
predicted gene 4876 |
1017 |
0.58 |
| chr4_150280961_150281112 | 0.17 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
610 |
0.68 |
| chr8_56233300_56233562 | 0.17 |
Hpgd |
hydroxyprostaglandin dehydrogenase 15 (NAD) |
61154 |
0.13 |
| chr4_63271316_63271493 | 0.17 |
Col27a1 |
collagen, type XXVII, alpha 1 |
11263 |
0.16 |
| chr16_78631922_78632084 | 0.17 |
Gm24861 |
predicted gene, 24861 |
7256 |
0.22 |
| chrX_11018346_11018497 | 0.17 |
Gm14485 |
predicted gene 14485 |
2586 |
0.39 |
| chr9_32005362_32005542 | 0.17 |
Gm47465 |
predicted gene, 47465 |
8 |
0.97 |
| chr3_24782580_24782752 | 0.17 |
Naaladl2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
956 |
0.73 |
| chr3_104220440_104221564 | 0.17 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
628 |
0.6 |
| chrX_52147782_52148137 | 0.17 |
Gpc4 |
glypican 4 |
17293 |
0.27 |
| chr11_101585925_101586098 | 0.17 |
Tmem106a |
transmembrane protein 106A |
14 |
0.94 |
| chr10_95254224_95254433 | 0.17 |
Gm48880 |
predicted gene, 48880 |
60525 |
0.08 |
| chr9_74894770_74895613 | 0.17 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr4_49435481_49435755 | 0.17 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
15516 |
0.12 |
| chr2_132638397_132638559 | 0.17 |
AU019990 |
expressed sequence AU019990 |
7403 |
0.13 |
| chr7_5071633_5071806 | 0.16 |
U2af2 |
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 2 |
3689 |
0.08 |
| chr7_59241435_59241594 | 0.16 |
Ube3a |
ubiquitin protein ligase E3A |
1394 |
0.16 |
| chr11_16883025_16883203 | 0.16 |
Egfr |
epidermal growth factor receptor |
4964 |
0.23 |
| chr12_119071015_119071241 | 0.16 |
Gm18921 |
predicted gene, 18921 |
3698 |
0.34 |
| chr15_9602541_9602716 | 0.16 |
Il7r |
interleukin 7 receptor |
72452 |
0.11 |
| chr12_78228003_78228154 | 0.16 |
Gphn |
gephyrin |
1425 |
0.29 |
| chr2_84386945_84387393 | 0.16 |
Calcrl |
calcitonin receptor-like |
11811 |
0.2 |
| chr1_169166546_169166738 | 0.16 |
Mir6354 |
microRNA 6354 |
147553 |
0.04 |
| chr11_16885540_16885691 | 0.16 |
Egfr |
epidermal growth factor receptor |
7465 |
0.21 |
| chr4_131041255_131041582 | 0.16 |
A930031H19Rik |
RIKEN cDNA A930031H19 gene |
30944 |
0.18 |
| chr9_122807315_122807480 | 0.16 |
Tcaim |
T cell activation inhibitor, mitochondrial |
1794 |
0.25 |
| chr9_61163963_61164339 | 0.16 |
I730028E13Rik |
RIKEN cDNA I730028E13 gene |
25636 |
0.14 |
| chr11_44683144_44683318 | 0.16 |
Gm12158 |
predicted gene 12158 |
20555 |
0.2 |
| chr4_57123440_57123604 | 0.16 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
3462 |
0.28 |
| chrX_51399106_51399311 | 0.16 |
Gm14621 |
predicted gene 14621 |
159676 |
0.03 |
| chr4_142221610_142221783 | 0.16 |
Kazn |
kazrin, periplakin interacting protein |
11490 |
0.22 |
| chr3_16184013_16184168 | 0.15 |
Ythdf3 |
YTH N6-methyladenosine RNA binding protein 3 |
854 |
0.71 |
| chr6_141856209_141856370 | 0.15 |
Slco1a4 |
solute carrier organic anion transporter family, member 1a4 |
90 |
0.97 |
| chr8_105045983_105046145 | 0.15 |
Gm45725 |
predicted gene 45725 |
898 |
0.39 |
| chr6_71193278_71193676 | 0.15 |
Fabp1 |
fatty acid binding protein 1, liver |
6350 |
0.13 |
| chr5_146224346_146224522 | 0.15 |
Rnf6 |
ring finger protein (C3H2C3 type) 6 |
2879 |
0.15 |
| chr4_84540692_84540849 | 0.15 |
Bnc2 |
basonuclin 2 |
5520 |
0.32 |
| chr9_109126916_109127177 | 0.15 |
Plxnb1 |
plexin B1 |
20252 |
0.09 |
| chr1_50955186_50955338 | 0.15 |
Tmeff2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
27743 |
0.19 |
| chr4_3402550_3402701 | 0.15 |
Gm11784 |
predicted gene 11784 |
15779 |
0.19 |
| chr7_102184013_102184287 | 0.15 |
Nup98 |
nucleoporin 98 |
585 |
0.66 |
| chr13_44339412_44339567 | 0.15 |
Gm33958 |
predicted gene, 33958 |
51628 |
0.11 |
| chr11_45255693_45255855 | 0.15 |
Gm12161 |
predicted gene 12161 |
25180 |
0.18 |
| chr7_100367751_100367915 | 0.15 |
Ppme1 |
protein phosphatase methylesterase 1 |
245 |
0.89 |
| chr9_37082260_37082465 | 0.15 |
Pknox2 |
Pbx/knotted 1 homeobox 2 |
917 |
0.53 |
| chrX_108664177_108664328 | 0.14 |
Gm379 |
predicted gene 379 |
203 |
0.97 |
| chr5_105353588_105353739 | 0.14 |
Gbp11 |
guanylate binding protein 11 |
7191 |
0.18 |
| chrX_163768401_163768583 | 0.14 |
Rnf138rt1 |
ring finger protein 138, retrogene 1 |
7163 |
0.23 |
| chr1_75342981_75343140 | 0.14 |
Des |
desmin |
17269 |
0.09 |
| chr18_81321681_81321954 | 0.14 |
Gm30192 |
predicted gene, 30192 |
57109 |
0.13 |
| chr2_94066434_94066607 | 0.14 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
51 |
0.98 |
| chr8_41473406_41473602 | 0.14 |
Frg1 |
FSHD region gene 1 |
56430 |
0.14 |
| chr19_20723027_20723214 | 0.14 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
4442 |
0.3 |
| chr16_43230215_43230536 | 0.14 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
5505 |
0.24 |
| chr2_109955276_109955427 | 0.14 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
20191 |
0.18 |
| chr14_98816721_98816872 | 0.14 |
Gm27034 |
predicted gene, 27034 |
101983 |
0.06 |
| chr5_36563902_36564053 | 0.14 |
Tbc1d14 |
TBC1 domain family, member 14 |
79 |
0.97 |
| chrX_57010435_57010605 | 0.14 |
Brs3 |
bombesin-like receptor 3 |
32554 |
0.15 |
| chr17_21534528_21534684 | 0.14 |
Zfp52 |
zinc finger protein 52 |
933 |
0.35 |
| chr8_5030377_5030626 | 0.14 |
n-R5s93 |
nuclear encoded rRNA 5S 93 |
38876 |
0.14 |
| chr1_4768744_4768905 | 0.14 |
Gm6123 |
predicted gene 6123 |
2307 |
0.18 |
| chr10_112926082_112926233 | 0.14 |
Gm26596 |
predicted gene, 26596 |
2344 |
0.23 |
| chr16_43532035_43532200 | 0.14 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
21809 |
0.21 |
| chr19_30241662_30241818 | 0.14 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
8798 |
0.21 |
| chr2_52576390_52576567 | 0.14 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
17911 |
0.18 |
| chr15_77151879_77152047 | 0.14 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
1593 |
0.31 |
| chr3_98682461_98682815 | 0.13 |
Gm12399 |
predicted gene 12399 |
4307 |
0.16 |
| chr15_38712864_38713163 | 0.13 |
Gm45924 |
predicted gene, 45924 |
15503 |
0.18 |
| chr7_69980197_69980349 | 0.13 |
Gm24120 |
predicted gene, 24120 |
22369 |
0.2 |
| chr2_67888984_67889171 | 0.13 |
Gm37964 |
predicted gene, 37964 |
9519 |
0.26 |
| chr3_142777238_142777410 | 0.13 |
Gm24457 |
predicted gene, 24457 |
4248 |
0.14 |
| chr11_93988000_93988159 | 0.13 |
Spag9 |
sperm associated antigen 9 |
8012 |
0.15 |
| chrX_38452345_38452518 | 0.13 |
Lamp2 |
lysosomal-associated membrane protein 2 |
3983 |
0.21 |
| chr1_164415069_164415280 | 0.13 |
Gm37411 |
predicted gene, 37411 |
9539 |
0.16 |
| chr6_115465552_115465760 | 0.13 |
Gm44079 |
predicted gene, 44079 |
666 |
0.7 |
| chr8_94594314_94594528 | 0.13 |
Fam192a |
family with sequence similarity 192, member A |
7130 |
0.12 |
| chr3_80042843_80043016 | 0.13 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
11963 |
0.22 |
| chr7_118250628_118250799 | 0.13 |
4930583K01Rik |
RIKEN cDNA 4930583K01 gene |
6848 |
0.14 |
| chr3_82036127_82036278 | 0.13 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
880 |
0.6 |
| chr10_87931882_87932033 | 0.13 |
Tyms-ps |
thymidylate synthase, pseudogene |
34890 |
0.14 |
| chr17_91875114_91875285 | 0.13 |
Gm41654 |
predicted gene, 41654 |
3060 |
0.27 |
| chr5_96980131_96980330 | 0.13 |
Gm9484 |
predicted gene 9484 |
17134 |
0.11 |
| chr4_105655884_105656063 | 0.13 |
Gm12726 |
predicted gene 12726 |
77633 |
0.11 |
| chr13_64106036_64106228 | 0.13 |
Slc35d2 |
solute carrier family 35, member D2 |
1031 |
0.46 |
| chr13_50815744_50815906 | 0.13 |
Gm2654 |
predicted gene 2654 |
29681 |
0.16 |
| chr15_3608242_3608402 | 0.13 |
Gm22031 |
predicted gene, 22031 |
4925 |
0.27 |
| chr6_48642896_48643049 | 0.13 |
Gimap8 |
GTPase, IMAP family member 8 |
4262 |
0.08 |
| chr14_27673339_27673537 | 0.13 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
20611 |
0.21 |
| chr3_104273850_104274019 | 0.13 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
53560 |
0.08 |
| chr16_57127831_57128340 | 0.13 |
Tomm70a |
translocase of outer mitochondrial membrane 70A |
6382 |
0.18 |
| chr16_46833812_46833992 | 0.13 |
Gm6912 |
predicted gene 6912 |
240296 |
0.02 |
| chr19_3194323_3194505 | 0.13 |
1700030N03Rik |
RIKEN cDNA 1700030N03 gene |
3284 |
0.14 |
| chrX_85452459_85452626 | 0.13 |
Gm14765 |
predicted gene 14765 |
20715 |
0.24 |
| chr16_42836264_42836446 | 0.13 |
4932412D23Rik |
RIKEN cDNA 4932412D23 gene |
39232 |
0.16 |
| chr13_31531112_31531384 | 0.13 |
Foxq1 |
forkhead box Q1 |
24886 |
0.13 |
| chr12_85785468_85785654 | 0.13 |
Flvcr2 |
feline leukemia virus subgroup C cellular receptor 2 |
10731 |
0.15 |
| chr11_16791951_16792316 | 0.13 |
Egfros |
epidermal growth factor receptor, opposite strand |
38569 |
0.14 |
| chr13_54665182_54665344 | 0.13 |
Faf2 |
Fas associated factor family member 2 |
13822 |
0.12 |
| chr11_16798476_16798627 | 0.13 |
Egfros |
epidermal growth factor receptor, opposite strand |
32151 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.0 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |