Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
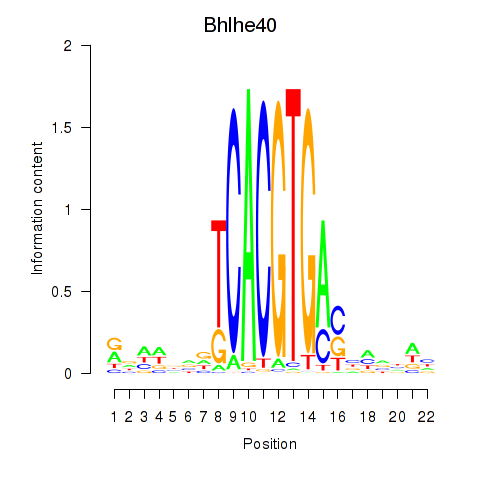
Results for Bhlhe40
Z-value: 1.50
Transcription factors associated with Bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bhlhe40
|
ENSMUSG00000030103.5 | basic helix-loop-helix family, member e40 |
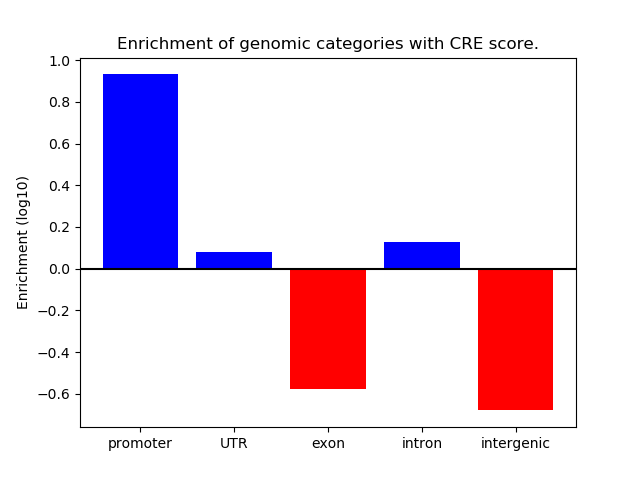
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_108709314_108709465 | Bhlhe40 | 46343 | 0.116291 | 0.73 | 9.9e-02 | Click! |
| chr6_108668460_108668647 | Bhlhe40 | 5507 | 0.177493 | -0.71 | 1.2e-01 | Click! |
| chr6_108704850_108705008 | Bhlhe40 | 41883 | 0.123574 | -0.69 | 1.3e-01 | Click! |
| chr6_108670913_108671087 | Bhlhe40 | 7954 | 0.168584 | -0.65 | 1.6e-01 | Click! |
| chr6_108664929_108665106 | Bhlhe40 | 1971 | 0.278122 | 0.65 | 1.6e-01 | Click! |
Activity of the Bhlhe40 motif across conditions
Conditions sorted by the z-value of the Bhlhe40 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
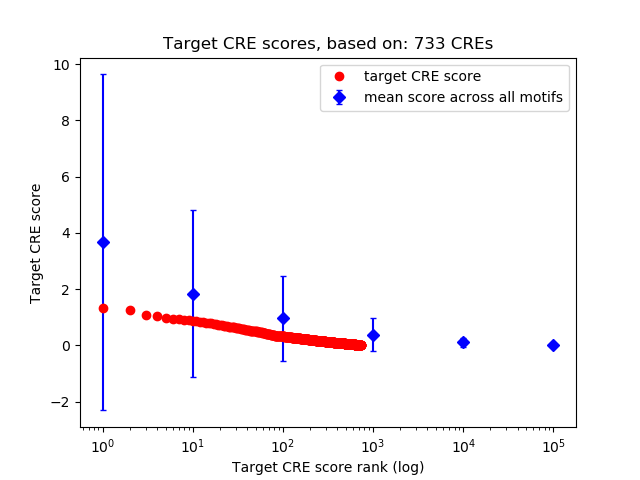
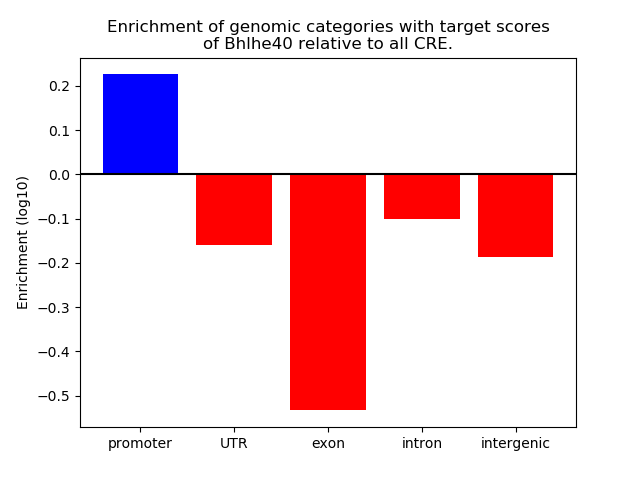
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_93323421_93323587 | 1.31 |
Ccng2 |
cyclin G2 |
54796 |
0.1 |
| chr5_25015695_25015994 | 1.25 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
20091 |
0.17 |
| chr19_4201629_4201786 | 1.07 |
Rad9a |
RAD9 checkpoint clamp component A |
45 |
0.5 |
| chr19_7240751_7240928 | 1.03 |
Naa40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
195 |
0.89 |
| chr7_81002031_81002182 | 0.97 |
Gm44967 |
predicted gene 44967 |
7423 |
0.11 |
| chr19_23074772_23074923 | 0.95 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
1006 |
0.54 |
| chr3_121735675_121736039 | 0.95 |
F3 |
coagulation factor III |
6310 |
0.11 |
| chr4_149426608_149426780 | 0.91 |
Ube4b |
ubiquitination factor E4B |
55 |
0.96 |
| chr2_103592357_103592552 | 0.90 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
26144 |
0.17 |
| chr13_38849595_38849761 | 0.86 |
Gm46392 |
predicted gene, 46392 |
12052 |
0.18 |
| chr6_115838132_115838303 | 0.85 |
Efcab12 |
EF-hand calcium binding domain 12 |
195 |
0.88 |
| chr5_136170628_136170833 | 0.82 |
Orai2 |
ORAI calcium release-activated calcium modulator 2 |
17 |
0.96 |
| chr17_24205766_24206106 | 0.81 |
Tbc1d24 |
TBC1 domain family, member 24 |
374 |
0.68 |
| chr9_44079482_44079643 | 0.81 |
Usp2 |
ubiquitin specific peptidase 2 |
5377 |
0.08 |
| chr5_115010965_115011126 | 0.80 |
Sppl3 |
signal peptide peptidase 3 |
92 |
0.94 |
| chr17_12769661_12769836 | 0.78 |
Igf2r |
insulin-like growth factor 2 receptor |
84 |
0.95 |
| chr10_43742623_43743061 | 0.76 |
Gm40634 |
predicted gene, 40634 |
245 |
0.89 |
| chr9_49365685_49366095 | 0.75 |
Drd2 |
dopamine receptor D2 |
25263 |
0.2 |
| chr1_143739744_143740319 | 0.73 |
Glrx2 |
glutaredoxin 2 (thioltransferase) |
344 |
0.84 |
| chr11_117780625_117780787 | 0.72 |
Tmc6 |
transmembrane channel-like gene family 6 |
48 |
0.93 |
| chr19_30096271_30096642 | 0.71 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
4495 |
0.24 |
| chr15_58972603_58972890 | 0.70 |
Mtss1 |
MTSS I-BAR domain containing 1 |
207 |
0.92 |
| chr6_50588087_50588294 | 0.69 |
4921507P07Rik |
RIKEN cDNA 4921507P07 gene |
8442 |
0.09 |
| chr7_45466827_45467004 | 0.68 |
Bax |
BCL2-associated X protein |
17 |
0.91 |
| chr1_89940488_89940663 | 0.67 |
Gbx2 |
gastrulation brain homeobox 2 |
9396 |
0.19 |
| chr6_9078648_9078799 | 0.66 |
Gm35736 |
predicted gene, 35736 |
68043 |
0.14 |
| chr1_39102289_39102448 | 0.65 |
Gm37091 |
predicted gene, 37091 |
19030 |
0.18 |
| chr3_67582623_67582799 | 0.64 |
Mfsd1 |
major facilitator superfamily domain containing 1 |
30 |
0.96 |
| chr5_104046470_104046646 | 0.64 |
Nudt9 |
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
32 |
0.92 |
| chrX_134600952_134601288 | 0.63 |
Gla |
galactosidase, alpha |
5 |
0.52 |
| chr11_120493774_120493943 | 0.62 |
Gm11788 |
predicted gene 11788 |
1464 |
0.16 |
| chr8_11730299_11730450 | 0.62 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
2197 |
0.17 |
| chr18_12168562_12168722 | 0.61 |
Rmc1 |
regulator of MON1-CCZ1 |
75 |
0.96 |
| chr7_46795972_46796137 | 0.60 |
Hps5 |
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
10 |
0.51 |
| chr13_102013412_102013573 | 0.58 |
Gm17832 |
predicted gene, 17832 |
93072 |
0.08 |
| chr8_124569182_124569523 | 0.57 |
Agt |
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
354 |
0.86 |
| chr9_40638323_40638501 | 0.56 |
Gm48284 |
predicted gene, 48284 |
22026 |
0.1 |
| chr10_40371945_40372096 | 0.56 |
Cdk19os |
cyclin-dependent kinase 19, opposite strand |
21917 |
0.13 |
| chr2_22998932_22999118 | 0.56 |
Gm13357 |
predicted gene 13357 |
20642 |
0.15 |
| chr6_121886502_121886729 | 0.53 |
Mug1 |
murinoglobulin 1 |
1038 |
0.53 |
| chr1_21261382_21261702 | 0.53 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8021 |
0.11 |
| chr4_118620306_118620678 | 0.53 |
Cfap57 |
cilia and flagella associated protein 57 |
87 |
0.76 |
| chr8_70766641_70767079 | 0.52 |
Ifi30 |
interferon gamma inducible protein 30 |
197 |
0.55 |
| chr17_28517484_28517972 | 0.52 |
Fkbp5 |
FK506 binding protein 5 |
201 |
0.84 |
| chr18_32287263_32287414 | 0.52 |
Ercc3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
30069 |
0.16 |
| chr9_21367416_21367785 | 0.52 |
Ilf3 |
interleukin enhancer binding factor 3 |
271 |
0.81 |
| chr9_35125828_35126176 | 0.51 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
6734 |
0.13 |
| chr7_55911807_55911995 | 0.51 |
Cyfip1 |
cytoplasmic FMR1 interacting protein 1 |
3529 |
0.15 |
| chr18_76089817_76090552 | 0.50 |
2900057B20Rik |
RIKEN cDNA 2900057B20 gene |
8287 |
0.21 |
| chr14_57746066_57746227 | 0.49 |
Lats2 |
large tumor suppressor 2 |
23 |
0.51 |
| chr10_128923444_128923601 | 0.49 |
Bloc1s1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
2 |
0.9 |
| chr2_144369139_144369688 | 0.49 |
Kat14 |
lysine acetyltransferase 14 |
414 |
0.43 |
| chr5_90338287_90338461 | 0.48 |
Ankrd17 |
ankyrin repeat domain 17 |
1369 |
0.45 |
| chr19_41560568_41560719 | 0.47 |
Lcor |
ligand dependent nuclear receptor corepressor |
11004 |
0.2 |
| chr9_107618752_107619076 | 0.47 |
Gnai2 |
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
1442 |
0.16 |
| chr14_79488949_79489435 | 0.47 |
Wbp4 |
WW domain binding protein 4 |
7672 |
0.16 |
| chr10_75510858_75511147 | 0.46 |
Gucd1 |
guanylyl cyclase domain containing 1 |
262 |
0.86 |
| chr16_35769456_35769643 | 0.46 |
Slc49a4 |
solute carrier family 49 member 4 |
193 |
0.86 |
| chr19_54115140_54115291 | 0.45 |
Gm50186 |
predicted gene, 50186 |
30656 |
0.18 |
| chr13_18121582_18121957 | 0.45 |
4930448F12Rik |
RIKEN cDNA 4930448F12 gene |
21567 |
0.15 |
| chr5_66042796_66042956 | 0.44 |
Rbm47 |
RNA binding motif protein 47 |
11676 |
0.12 |
| chr11_52000637_52000943 | 0.44 |
Ube2b |
ubiquitin-conjugating enzyme E2B |
28 |
0.52 |
| chr10_4334368_4334532 | 0.42 |
Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
95 |
0.97 |
| chr14_50924662_50925001 | 0.42 |
Osgep |
O-sialoglycoprotein endopeptidase |
1 |
0.62 |
| chr5_74401642_74401799 | 0.41 |
Scfd2 |
Sec1 family domain containing 2 |
3982 |
0.2 |
| chr3_153906036_153906187 | 0.41 |
Msh4 |
mutS homolog 4 |
27 |
0.94 |
| chr19_5610162_5610532 | 0.41 |
Kat5 |
K(lysine) acetyltransferase 5 |
110 |
0.91 |
| chr10_77101414_77101603 | 0.41 |
Col18a1 |
collagen, type XVIII, alpha 1 |
12080 |
0.17 |
| chr13_46735806_46736396 | 0.41 |
Nup153 |
nucleoporin 153 |
8161 |
0.17 |
| chr5_103778320_103778486 | 0.40 |
Aff1 |
AF4/FMR2 family, member 1 |
5956 |
0.23 |
| chr17_44350675_44350826 | 0.39 |
Gm49872 |
predicted gene, 49872 |
17153 |
0.27 |
| chr10_22731916_22732212 | 0.39 |
Tbpl1 |
TATA box binding protein-like 1 |
126 |
0.9 |
| chr15_80255398_80255563 | 0.38 |
Atf4 |
activating transcription factor 4 |
43 |
0.95 |
| chr19_55593151_55593306 | 0.37 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
148592 |
0.04 |
| chr7_5062430_5062624 | 0.37 |
U2af2 |
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 2 |
223 |
0.65 |
| chr2_84742449_84742615 | 0.37 |
Gm19426 |
predicted gene, 19426 |
1123 |
0.24 |
| chr17_22361357_22361742 | 0.36 |
Zfp758 |
zinc finger protein 758 |
27 |
0.65 |
| chr10_61143381_61143586 | 0.36 |
Sgpl1 |
sphingosine phosphate lyase 1 |
3415 |
0.19 |
| chr5_116419061_116419437 | 0.35 |
Hspb8 |
heat shock protein 8 |
3357 |
0.16 |
| chr15_97285773_97285959 | 0.35 |
Gm4383 |
predicted gene 4383 |
17108 |
0.2 |
| chr9_77961400_77961736 | 0.35 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
20277 |
0.12 |
| chr13_111532342_111532493 | 0.35 |
Gm15290 |
predicted gene 15290 |
17494 |
0.13 |
| chr15_77520533_77520827 | 0.34 |
Apol11a |
apolipoprotein L 11a |
12409 |
0.11 |
| chr1_132586686_132587343 | 0.34 |
Nfasc |
neurofascin |
126 |
0.97 |
| chr4_127077236_127077665 | 0.34 |
Zmym6 |
zinc finger, MYM-type 6 |
8 |
0.96 |
| chr1_13171208_13171385 | 0.34 |
Gm38376 |
predicted gene, 38376 |
8418 |
0.16 |
| chr12_84222439_84222605 | 0.34 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
3641 |
0.14 |
| chr18_34572683_34572834 | 0.34 |
Nme5 |
NME/NM23 family member 5 |
6325 |
0.14 |
| chr10_128322189_128322510 | 0.34 |
Cnpy2 |
canopy FGF signaling regulator 2 |
110 |
0.9 |
| chr1_40085784_40085959 | 0.34 |
Gm16894 |
predicted gene, 16894 |
20 |
0.6 |
| chr2_130424229_130424389 | 0.33 |
Pced1a |
PC-esterase domain containing 1A |
1 |
0.5 |
| chr14_30714856_30715239 | 0.33 |
Sfmbt1 |
Scm-like with four mbt domains 1 |
120 |
0.96 |
| chr11_55204221_55204382 | 0.33 |
Slc36a1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
49 |
0.97 |
| chr6_34176282_34176719 | 0.33 |
Slc35b4 |
solute carrier family 35, member B4 |
470 |
0.78 |
| chr8_105773166_105773381 | 0.33 |
Gfod2 |
glucose-fructose oxidoreductase domain containing 2 |
14609 |
0.1 |
| chr6_91707022_91707254 | 0.32 |
Gm45828 |
predicted gene 45828 |
2268 |
0.2 |
| chr13_41001139_41001299 | 0.32 |
Pak1ip1 |
PAK1 interacting protein 1 |
19 |
0.53 |
| chr12_99208180_99208477 | 0.32 |
Gm19898 |
predicted gene, 19898 |
37343 |
0.14 |
| chr9_123717598_123717778 | 0.32 |
Lztfl1 |
leucine zipper transcription factor-like 1 |
9 |
0.96 |
| chr5_72241549_72241735 | 0.32 |
Atp10d |
ATPase, class V, type 10D |
23002 |
0.17 |
| chr7_90129909_90130310 | 0.32 |
Picalm |
phosphatidylinositol binding clathrin assembly protein |
104 |
0.52 |
| chr8_35707942_35708100 | 0.32 |
1700015I17Rik |
RIKEN cDNA 1700015I17 gene |
11928 |
0.15 |
| chr9_46036951_46037323 | 0.31 |
Sik3 |
SIK family kinase 3 |
24137 |
0.13 |
| chr14_41013764_41013924 | 0.31 |
Prxl2a |
peroxiredoxin like 2A |
56 |
0.97 |
| chr7_46795676_46795831 | 0.31 |
Hps5 |
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
50 |
0.81 |
| chr10_89490631_89490857 | 0.31 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
15905 |
0.21 |
| chr12_65036195_65036354 | 0.31 |
Prpf39 |
pre-mRNA processing factor 39 |
59 |
0.96 |
| chr9_43545212_43545661 | 0.31 |
Gm36855 |
predicted gene, 36855 |
24432 |
0.17 |
| chr3_116329378_116329892 | 0.30 |
Gm29151 |
predicted gene 29151 |
20468 |
0.17 |
| chr10_19015307_19015840 | 0.30 |
Tnfaip3 |
tumor necrosis factor, alpha-induced protein 3 |
84 |
0.98 |
| chr2_6338002_6338625 | 0.30 |
Usp6nl |
USP6 N-terminal like |
14420 |
0.18 |
| chr15_97018730_97018892 | 0.30 |
Slc38a4 |
solute carrier family 38, member 4 |
1140 |
0.62 |
| chr16_91255820_91256295 | 0.30 |
Gm37016 |
predicted gene, 37016 |
6441 |
0.13 |
| chr11_101553520_101553877 | 0.29 |
Nbr1 |
NBR1, autophagy cargo receptor |
797 |
0.4 |
| chr9_87255292_87255453 | 0.29 |
Cep162 |
centrosomal protein 162 |
84 |
0.98 |
| chr6_128362234_128363003 | 0.29 |
Rhno1 |
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
39 |
0.8 |
| chr7_81004740_81004957 | 0.29 |
Gm44967 |
predicted gene 44967 |
10165 |
0.11 |
| chr8_84792517_84792893 | 0.29 |
Nfix |
nuclear factor I/X |
7319 |
0.1 |
| chr6_94104151_94104302 | 0.29 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
178799 |
0.03 |
| chr7_35802959_35803128 | 0.29 |
Zfp507 |
zinc finger protein 507 |
40 |
0.88 |
| chr5_87086248_87086432 | 0.29 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
4817 |
0.13 |
| chr3_89202795_89202962 | 0.29 |
Gba |
glucosidase, beta, acid |
50 |
0.9 |
| chr13_85189268_85189450 | 0.28 |
Ccnh |
cyclin H |
49 |
0.98 |
| chr6_99523725_99523876 | 0.28 |
Foxp1 |
forkhead box P1 |
1079 |
0.57 |
| chr11_58033500_58033932 | 0.28 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
7897 |
0.15 |
| chr8_125669724_125669887 | 0.28 |
Map10 |
microtubule-associated protein 10 |
13 |
0.98 |
| chr4_135420119_135420277 | 0.28 |
Mir700 |
microRNA 700 |
3565 |
0.12 |
| chr4_8799595_8799756 | 0.28 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
40647 |
0.2 |
| chr5_150774025_150774397 | 0.28 |
Pds5b |
PDS5 cohesin associated factor B |
14847 |
0.15 |
| chr9_94053316_94053467 | 0.28 |
Gm5369 |
predicted gene 5369 |
86071 |
0.11 |
| chr12_102878478_102878658 | 0.27 |
Btbd7 |
BTB (POZ) domain containing 7 |
97 |
0.95 |
| chr9_22389111_22389373 | 0.27 |
Anln |
anillin, actin binding protein |
54 |
0.95 |
| chr17_66077011_66077179 | 0.27 |
Ankrd12 |
ankyrin repeat domain 12 |
6 |
0.96 |
| chr13_69531330_69531517 | 0.27 |
Tent4a |
terminal nucleotidyltransferase 4A |
1009 |
0.41 |
| chr3_81386912_81387263 | 0.27 |
Gm37300 |
predicted gene, 37300 |
18485 |
0.28 |
| chr7_19629444_19629613 | 0.27 |
Relb |
avian reticuloendotheliosis viral (v-rel) oncogene related B |
90 |
0.93 |
| chr2_36159845_36160023 | 0.27 |
Mrrf |
mitochondrial ribosome recycling factor |
12831 |
0.12 |
| chr17_56479539_56479705 | 0.27 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
3139 |
0.2 |
| chr5_124425319_124425853 | 0.27 |
Sbno1 |
strawberry notch 1 |
2 |
0.38 |
| chr19_9977949_9978176 | 0.27 |
Fth1 |
ferritin heavy polypeptide 1 |
2536 |
0.15 |
| chr2_166126070_166126225 | 0.26 |
Sulf2 |
sulfatase 2 |
28276 |
0.15 |
| chr6_21823423_21823592 | 0.26 |
Tspan12 |
tetraspanin 12 |
28321 |
0.16 |
| chr19_46661548_46661819 | 0.26 |
Gm36602 |
predicted gene, 36602 |
4443 |
0.14 |
| chr14_31172747_31173116 | 0.26 |
Nisch |
nischarin |
797 |
0.47 |
| chr1_64690615_64690946 | 0.26 |
Ccnyl1 |
cyclin Y-like 1 |
42 |
0.59 |
| chr11_70000432_70000600 | 0.26 |
Dvl2 |
dishevelled segment polarity protein 2 |
79 |
0.9 |
| chr16_45158641_45158816 | 0.26 |
Slc35a5 |
solute carrier family 35, member A5 |
22 |
0.52 |
| chr17_24163699_24163861 | 0.26 |
Amdhd2 |
amidohydrolase domain containing 2 |
14 |
0.94 |
| chr5_146792230_146792423 | 0.26 |
Usp12 |
ubiquitin specific peptidase 12 |
2680 |
0.19 |
| chr4_139109272_139109459 | 0.26 |
Micos10 |
mitochondrial contact site and cristae organizing system subunit 10 |
1878 |
0.25 |
| chr3_121214666_121215038 | 0.25 |
Tlcd4 |
TLC domain containing 4 |
16681 |
0.14 |
| chr16_92826008_92826183 | 0.25 |
Runx1 |
runt related transcription factor 1 |
21 |
0.98 |
| chr10_95685288_95685475 | 0.25 |
Anapc15-ps |
anaphase promoting complex C subunit 15, pseudogene |
11930 |
0.11 |
| chr17_32943503_32943656 | 0.25 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
3756 |
0.12 |
| chr5_87562697_87562848 | 0.24 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
2581 |
0.17 |
| chr17_65886042_65886193 | 0.24 |
Gm49870 |
predicted gene, 49870 |
224 |
0.43 |
| chr11_5917118_5917412 | 0.24 |
Gm11967 |
predicted gene 11967 |
1950 |
0.2 |
| chr6_6577877_6578251 | 0.24 |
Sem1 |
SEM1, 26S proteasome complex subunit |
594 |
0.79 |
| chr5_72259392_72259543 | 0.24 |
Atp10d |
ATPase, class V, type 10D |
5177 |
0.21 |
| chr1_130826570_130826742 | 0.23 |
Pigr |
polymeric immunoglobulin receptor |
28 |
0.96 |
| chr5_97090249_97090465 | 0.23 |
Paqr3 |
progestin and adipoQ receptor family member III |
7414 |
0.19 |
| chr16_97610441_97610883 | 0.23 |
Tmprss2 |
transmembrane protease, serine 2 |
307 |
0.91 |
| chr10_81176435_81176616 | 0.23 |
Eef2 |
eukaryotic translation elongation factor 2 |
106 |
0.89 |
| chr19_42779893_42780058 | 0.23 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
1 |
0.95 |
| chr2_25356295_25356463 | 0.23 |
Dpp7 |
dipeptidylpeptidase 7 |
20 |
0.93 |
| chr7_126583293_126583667 | 0.23 |
Cln3 |
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
43 |
0.92 |
| chr14_61680870_61681697 | 0.23 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr15_99723268_99723456 | 0.23 |
Gm16537 |
predicted gene 16537 |
1112 |
0.24 |
| chr14_63066975_63067226 | 0.23 |
Defb42 |
defensin beta 42 |
12027 |
0.11 |
| chr16_35770249_35770414 | 0.23 |
Hspbap1 |
Hspb associated protein 1 |
44 |
0.94 |
| chr3_95754384_95754717 | 0.23 |
Tars2 |
threonyl-tRNA synthetase 2, mitochondrial (putative) |
78 |
0.94 |
| chr4_138981088_138981239 | 0.23 |
Tmco4 |
transmembrane and coiled-coil domains 4 |
8254 |
0.16 |
| chr13_35036409_35036560 | 0.22 |
Gm48626 |
predicted gene, 48626 |
4538 |
0.14 |
| chr3_10439906_10440215 | 0.22 |
Snx16 |
sorting nexin 16 |
27 |
0.98 |
| chr5_143622347_143622510 | 0.22 |
Cyth3 |
cytohesin 3 |
19 |
0.98 |
| chr4_59254499_59254704 | 0.22 |
Gm12596 |
predicted gene 12596 |
5450 |
0.21 |
| chr7_27196105_27196414 | 0.22 |
Snrpa |
small nuclear ribonucleoprotein polypeptide A |
12 |
0.86 |
| chr9_107666466_107666787 | 0.22 |
Slc38a3 |
solute carrier family 38, member 3 |
748 |
0.41 |
| chr8_93234472_93234646 | 0.22 |
Ces1e |
carboxylesterase 1E |
4940 |
0.15 |
| chr2_11441861_11442106 | 0.22 |
Gm13296 |
predicted gene 13296 |
21759 |
0.11 |
| chr14_52197392_52197558 | 0.22 |
Gm26590 |
predicted gene, 26590 |
27 |
0.52 |
| chr1_106178120_106178315 | 0.22 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
6465 |
0.19 |
| chr8_11477778_11477940 | 0.21 |
E230013L22Rik |
RIKEN cDNA E230013L22 gene |
70 |
0.92 |
| chr18_6136125_6136303 | 0.21 |
Arhgap12 |
Rho GTPase activating protein 12 |
116 |
0.97 |
| chr8_48740269_48740425 | 0.21 |
Tenm3 |
teneurin transmembrane protein 3 |
65657 |
0.13 |
| chr11_78964690_78964876 | 0.21 |
Lgals9 |
lectin, galactose binding, soluble 9 |
5547 |
0.2 |
| chr2_30133618_30133793 | 0.21 |
Tbc1d13 |
TBC1 domain family, member 13 |
41 |
0.95 |
| chr14_25396821_25396972 | 0.21 |
Gm26660 |
predicted gene, 26660 |
17287 |
0.16 |
| chr13_93612829_93612980 | 0.21 |
Gm15622 |
predicted gene 15622 |
12478 |
0.16 |
| chr3_30792776_30792939 | 0.21 |
Sec62 |
SEC62 homolog (S. cerevisiae) |
18 |
0.93 |
| chr3_107278861_107279068 | 0.21 |
Lamtor5 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
35 |
0.97 |
| chr14_59200883_59201288 | 0.21 |
Rcbtb1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
124 |
0.96 |
| chr11_50431056_50431230 | 0.21 |
Rufy1 |
RUN and FYVE domain containing 1 |
18 |
0.98 |
| chr8_85025239_85025399 | 0.21 |
Asna1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
38 |
0.91 |
| chr12_28899893_28900063 | 0.21 |
Gm31508 |
predicted gene, 31508 |
10251 |
0.18 |
| chr2_152729731_152729890 | 0.20 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
6441 |
0.11 |
| chr4_134468257_134468415 | 0.20 |
Stmn1 |
stathmin 1 |
16 |
0.5 |
| chr14_40934609_40934760 | 0.20 |
Tspan14 |
tetraspanin 14 |
363 |
0.88 |
| chr3_152396303_152396479 | 0.20 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
259 |
0.84 |
| chr17_25240096_25240257 | 0.20 |
Tsr3 |
TSR3 20S rRNA accumulation |
6 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.7 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.2 | 0.5 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.6 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.2 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.2 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.3 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.4 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.6 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.9 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.0 | GO:0016414 | O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |