Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
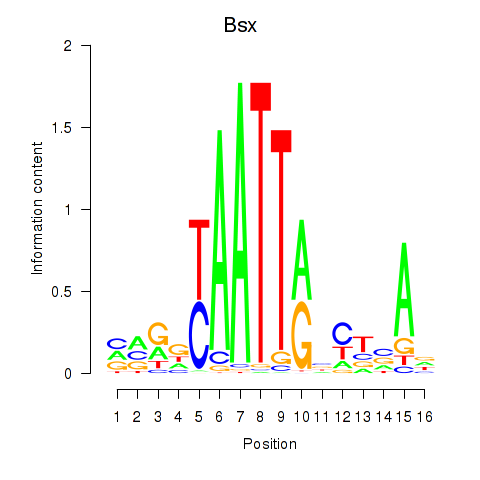
Results for Bsx
Z-value: 0.47
Transcription factors associated with Bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bsx
|
ENSMUSG00000054360.3 | brain specific homeobox |
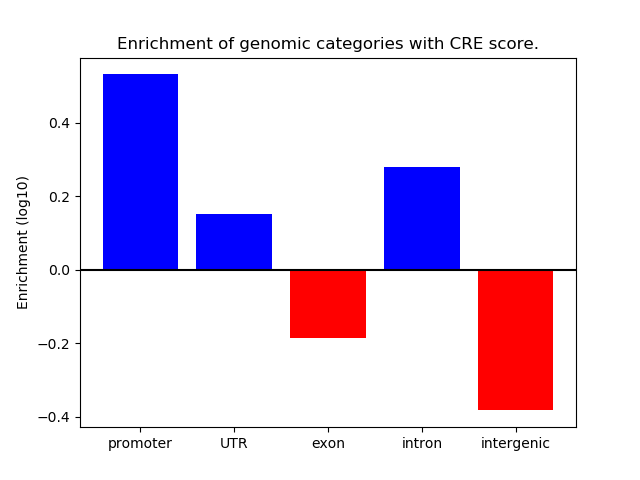
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_40879555_40879754 | Bsx | 5527 | 0.145007 | -0.33 | 5.3e-01 | Click! |
Activity of the Bsx motif across conditions
Conditions sorted by the z-value of the Bsx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
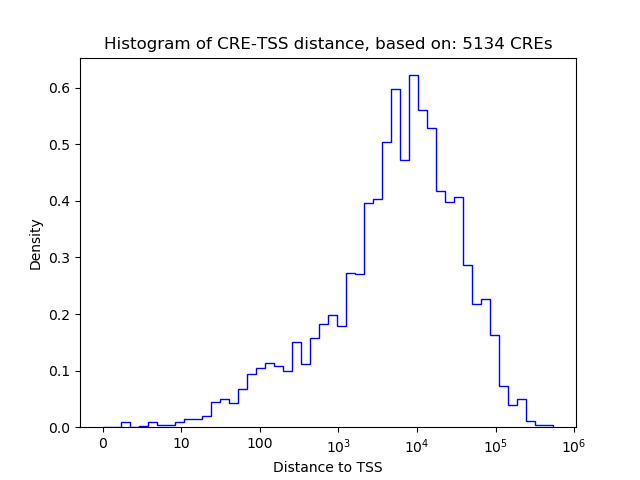
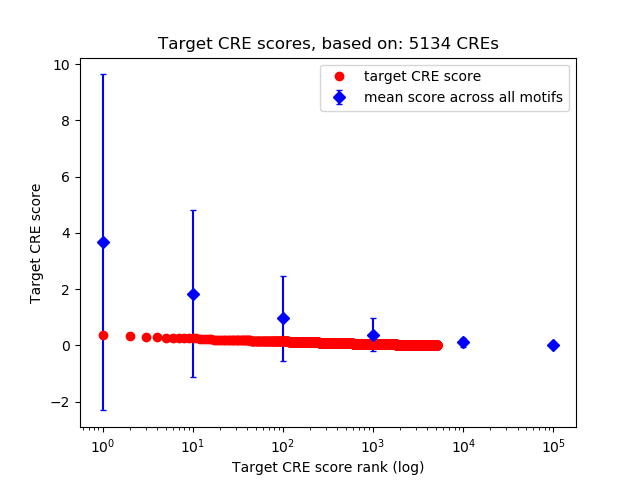
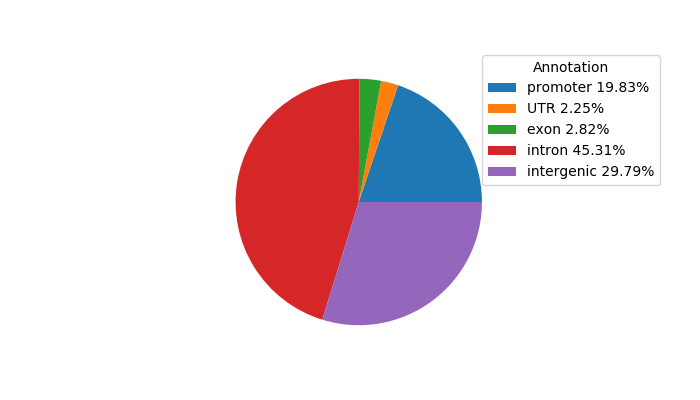
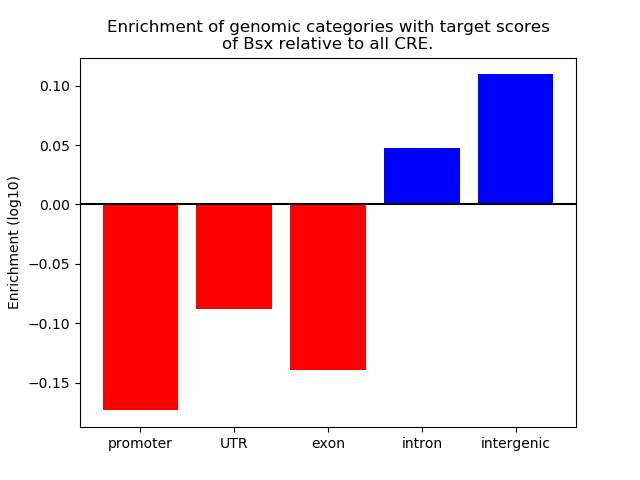
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_43971247_43971535 | 0.36 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr17_29123827_29123978 | 0.33 |
Rab44 |
RAB44, member RAS oncogene family |
9721 |
0.09 |
| chr12_84165735_84165922 | 0.29 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
5690 |
0.11 |
| chr13_80891058_80891263 | 0.28 |
Arrdc3 |
arrestin domain containing 3 |
642 |
0.68 |
| chr8_94172420_94173095 | 0.27 |
Mt2 |
metallothionein 2 |
93 |
0.88 |
| chr4_152989708_152989859 | 0.27 |
Gm25779 |
predicted gene, 25779 |
23801 |
0.26 |
| chr11_108936283_108936686 | 0.27 |
Axin2 |
axin 2 |
2390 |
0.3 |
| chr3_152143501_152143652 | 0.26 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
3212 |
0.17 |
| chr13_56593323_56593474 | 0.25 |
2010203P06Rik |
RIKEN cDNA 2010203P06 gene |
2139 |
0.29 |
| chr8_36668727_36668937 | 0.25 |
Dlc1 |
deleted in liver cancer 1 |
54889 |
0.16 |
| chr5_12476907_12477068 | 0.24 |
Gm8925 |
predicted gene 8925 |
2641 |
0.3 |
| chr2_126591150_126591329 | 0.22 |
Hdc |
histidine decarboxylase |
7022 |
0.18 |
| chr7_26881162_26881332 | 0.22 |
Cyp2a21-ps |
cytochrome P450, family 2, subfamily a, polypeptide 21, pseudogene |
36134 |
0.11 |
| chr2_22589828_22590052 | 0.21 |
Gm13340 |
predicted gene 13340 |
293 |
0.68 |
| chr15_93916844_93917358 | 0.21 |
9430014N10Rik |
RIKEN cDNA 9430014N10 gene |
10010 |
0.25 |
| chr2_50824872_50825029 | 0.21 |
Gm13498 |
predicted gene 13498 |
84734 |
0.1 |
| chr8_77961865_77962061 | 0.21 |
Gm29895 |
predicted gene, 29895 |
82047 |
0.09 |
| chr9_64774970_64775135 | 0.20 |
Dennd4a |
DENN/MADD domain containing 4A |
36288 |
0.13 |
| chr12_90731323_90731529 | 0.20 |
Gm26512 |
predicted gene, 26512 |
6775 |
0.21 |
| chr10_121611756_121611907 | 0.20 |
Xpot |
exportin, tRNA (nuclear export receptor for tRNAs) |
1762 |
0.27 |
| chr17_31323497_31323678 | 0.20 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
1333 |
0.36 |
| chr1_51513192_51513348 | 0.20 |
Gm17767 |
predicted gene, 17767 |
941 |
0.56 |
| chr2_7259683_7259857 | 0.20 |
Gm24340 |
predicted gene, 24340 |
90327 |
0.09 |
| chr12_84050784_84050944 | 0.20 |
Acot3 |
acyl-CoA thioesterase 3 |
1280 |
0.23 |
| chr16_87128308_87128459 | 0.20 |
Gm32865 |
predicted gene, 32865 |
56 |
0.99 |
| chr15_102150895_102151074 | 0.20 |
Soat2 |
sterol O-acyltransferase 2 |
132 |
0.92 |
| chr10_19585787_19585961 | 0.19 |
Ifngr1 |
interferon gamma receptor 1 |
6075 |
0.2 |
| chr7_145056434_145056606 | 0.19 |
Gm45181 |
predicted gene 45181 |
106476 |
0.05 |
| chr2_33444417_33444586 | 0.19 |
Gm13536 |
predicted gene 13536 |
2301 |
0.24 |
| chr18_7847563_7847725 | 0.19 |
Gm50027 |
predicted gene, 50027 |
2709 |
0.23 |
| chr17_5012388_5012845 | 0.19 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
16187 |
0.23 |
| chr19_32614009_32614160 | 0.19 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
5921 |
0.24 |
| chr13_45275232_45275389 | 0.19 |
Gm34466 |
predicted gene, 34466 |
7599 |
0.23 |
| chr5_27411249_27411405 | 0.18 |
Speer4b |
spermatogenesis associated glutamate (E)-rich protein 4B |
90035 |
0.09 |
| chr13_110577994_110578317 | 0.18 |
Gm33045 |
predicted gene, 33045 |
49133 |
0.17 |
| chr3_102976403_102976554 | 0.18 |
Nr1h5 |
nuclear receptor subfamily 1, group H, member 5 |
12345 |
0.12 |
| chr12_118887325_118887612 | 0.18 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
3291 |
0.3 |
| chr9_99502448_99502620 | 0.18 |
Nme9 |
NME/NM23 family member 9 |
32113 |
0.14 |
| chr2_75564827_75565114 | 0.18 |
Gm13655 |
predicted gene 13655 |
68412 |
0.08 |
| chr7_14430420_14431121 | 0.18 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
5677 |
0.17 |
| chr7_60003700_60003851 | 0.18 |
Snurf |
SNRPN upstream reading frame |
1274 |
0.17 |
| chr1_67124558_67125159 | 0.17 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
1832 |
0.42 |
| chr3_149934962_149935128 | 0.17 |
Rpsa-ps10 |
ribosomal protein SA, pseudogene 10 |
138497 |
0.05 |
| chr3_85831893_85832058 | 0.17 |
Fam160a1 |
family with sequence similarity 160, member A1 |
14684 |
0.15 |
| chr4_48315210_48315516 | 0.17 |
Gm12435 |
predicted gene 12435 |
29867 |
0.16 |
| chr2_57302912_57303068 | 0.17 |
Gm13535 |
predicted gene 13535 |
27304 |
0.16 |
| chr12_111449466_111449652 | 0.17 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
41 |
0.96 |
| chr19_43767883_43768034 | 0.17 |
Cutc |
cutC copper transporter |
3085 |
0.18 |
| chr3_41580995_41581174 | 0.17 |
Jade1 |
jade family PHD finger 1 |
228 |
0.92 |
| chr13_60267432_60267612 | 0.17 |
Gm24999 |
predicted gene, 24999 |
26150 |
0.16 |
| chr13_52976319_52976492 | 0.17 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
4668 |
0.2 |
| chr1_66862471_66862695 | 0.16 |
Acadl |
acyl-Coenzyme A dehydrogenase, long-chain |
694 |
0.5 |
| chr4_142367847_142368042 | 0.16 |
Gm13052 |
predicted gene 13052 |
21469 |
0.25 |
| chr8_115966051_115966449 | 0.16 |
Gm45733 |
predicted gene 45733 |
5821 |
0.34 |
| chr7_26087500_26087684 | 0.16 |
Gm29920 |
predicted gene, 29920 |
12724 |
0.13 |
| chr6_120479402_120479564 | 0.16 |
Il17ra |
interleukin 17 receptor A |
5880 |
0.14 |
| chr12_104772104_104772274 | 0.16 |
Clmn |
calmin |
8682 |
0.21 |
| chr4_150598739_150598957 | 0.16 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
16567 |
0.18 |
| chr1_118458813_118459291 | 0.16 |
Gm22710 |
predicted gene, 22710 |
213 |
0.44 |
| chr6_121890040_121890777 | 0.16 |
Mug1 |
murinoglobulin 1 |
4831 |
0.21 |
| chr7_122533976_122534175 | 0.16 |
Gm14389 |
predicted gene 14389 |
49655 |
0.11 |
| chr17_32382711_32382869 | 0.16 |
Wiz |
widely-interspaced zinc finger motifs |
4898 |
0.12 |
| chr3_25017765_25017942 | 0.16 |
Gm42774 |
predicted gene 42774 |
10148 |
0.29 |
| chr19_44027305_44027461 | 0.16 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
1816 |
0.28 |
| chr8_40869347_40869543 | 0.16 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
5503 |
0.17 |
| chr6_28704741_28704911 | 0.16 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
83 |
0.98 |
| chr7_72234078_72234229 | 0.16 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
4793 |
0.31 |
| chr1_72226394_72226550 | 0.16 |
Gm25360 |
predicted gene, 25360 |
232 |
0.89 |
| chr2_146865856_146866036 | 0.16 |
Kiz |
kizuna centrosomal protein |
10037 |
0.23 |
| chr7_102272655_102272818 | 0.16 |
Stim1 |
stromal interaction molecule 1 |
4409 |
0.15 |
| chr1_88121639_88121806 | 0.15 |
Gm15372 |
predicted gene 15372 |
5054 |
0.07 |
| chr1_59504281_59504519 | 0.15 |
Gm26813 |
predicted gene, 26813 |
11795 |
0.13 |
| chr5_63897626_63897962 | 0.15 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
20318 |
0.15 |
| chr12_73147698_73147884 | 0.15 |
Gm23910 |
predicted gene, 23910 |
1886 |
0.33 |
| chrX_157419778_157419945 | 0.15 |
Phex |
phosphate regulating endopeptidase homolog, X-linked |
4549 |
0.16 |
| chr1_153689607_153689785 | 0.15 |
Gm29529 |
predicted gene 29529 |
2960 |
0.17 |
| chr14_61045882_61046048 | 0.15 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
504 |
0.81 |
| chr2_97089216_97089367 | 0.15 |
4930445B16Rik |
RIKEN cDNA 4930445B16 gene |
5730 |
0.23 |
| chr2_160619907_160620070 | 0.15 |
Gm14221 |
predicted gene 14221 |
17 |
0.97 |
| chr6_121303535_121303719 | 0.15 |
Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
2845 |
0.19 |
| chr7_3145683_3146014 | 0.15 |
Gm29708 |
predicted gene, 29708 |
4143 |
0.09 |
| chr1_24613213_24613480 | 0.15 |
Gm28438 |
predicted gene 28438 |
227 |
0.6 |
| chr10_94565775_94566229 | 0.15 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
9255 |
0.17 |
| chr18_75180173_75180522 | 0.15 |
Gm8807 |
predicted gene 8807 |
2467 |
0.32 |
| chrX_18883144_18883317 | 0.15 |
Gm6923 |
predicted pseudogene 6923 |
37574 |
0.2 |
| chr7_140776294_140776445 | 0.15 |
Gm29799 |
predicted gene, 29799 |
8146 |
0.09 |
| chr3_96559546_96560080 | 0.15 |
Txnip |
thioredoxin interacting protein |
90 |
0.91 |
| chr7_115625366_115625533 | 0.15 |
Sox6 |
SRY (sex determining region Y)-box 6 |
27553 |
0.26 |
| chr1_40680680_40680831 | 0.15 |
Slc9a2 |
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
181 |
0.95 |
| chr18_65837283_65837437 | 0.14 |
Gm50174 |
predicted gene, 50174 |
73 |
0.94 |
| chr14_22964972_22965155 | 0.14 |
Gm47601 |
predicted gene, 47601 |
37137 |
0.22 |
| chr3_98682461_98682815 | 0.14 |
Gm12399 |
predicted gene 12399 |
4307 |
0.16 |
| chr1_21131249_21131442 | 0.14 |
Gm2693 |
predicted gene 2693 |
47643 |
0.1 |
| chr19_10168393_10168709 | 0.14 |
Gm50359 |
predicted gene, 50359 |
7610 |
0.11 |
| chr5_145222977_145223149 | 0.14 |
Zfp655 |
zinc finger protein 655 |
8652 |
0.1 |
| chr15_27993720_27993871 | 0.14 |
Trio |
triple functional domain (PTPRF interacting) |
1568 |
0.45 |
| chr12_80833658_80833843 | 0.14 |
Susd6 |
sushi domain containing 6 |
34958 |
0.11 |
| chr2_22587937_22588311 | 0.14 |
Gm13341 |
predicted gene 13341 |
162 |
0.91 |
| chr3_81386912_81387263 | 0.14 |
Gm37300 |
predicted gene, 37300 |
18485 |
0.28 |
| chr1_89670472_89670636 | 0.14 |
4933400F21Rik |
RIKEN cDNA 4933400F21 gene |
13334 |
0.24 |
| chr17_4004285_4004445 | 0.14 |
4930470H14Rik |
RIKEN cDNA 4930470H14 gene |
78630 |
0.11 |
| chr14_45050200_45050422 | 0.14 |
Gm21010 |
predicted gene, 21010 |
40304 |
0.08 |
| chr14_122862763_122862933 | 0.14 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
13087 |
0.2 |
| chr10_87453108_87453284 | 0.14 |
Ascl1 |
achaete-scute family bHLH transcription factor 1 |
40464 |
0.15 |
| chr1_152032225_152032416 | 0.14 |
1700025G04Rik |
RIKEN cDNA 1700025G04 gene |
22972 |
0.23 |
| chr19_57321939_57322277 | 0.14 |
Gm50270 |
predicted gene, 50270 |
1494 |
0.35 |
| chr11_116470132_116470520 | 0.14 |
Prpsap1 |
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
2917 |
0.13 |
| chr2_118318601_118318758 | 0.14 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
13765 |
0.13 |
| chr3_85550361_85550689 | 0.14 |
Gm42812 |
predicted gene 42812 |
2864 |
0.24 |
| chr7_90335926_90336077 | 0.14 |
Sytl2 |
synaptotagmin-like 2 |
12697 |
0.17 |
| chr15_31279226_31279377 | 0.14 |
Gm49296 |
predicted gene, 49296 |
2788 |
0.19 |
| chr10_80758685_80759184 | 0.14 |
Dot1l |
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
3471 |
0.11 |
| chr8_36618215_36618392 | 0.14 |
Dlc1 |
deleted in liver cancer 1 |
4360 |
0.32 |
| chr4_150685463_150685649 | 0.14 |
Gm16079 |
predicted gene 16079 |
6764 |
0.21 |
| chr5_105698740_105698909 | 0.14 |
Lrrc8d |
leucine rich repeat containing 8D |
1145 |
0.57 |
| chr1_170956684_170956835 | 0.13 |
Fcgr2b |
Fc receptor, IgG, low affinity IIb |
4871 |
0.1 |
| chr3_142777238_142777410 | 0.13 |
Gm24457 |
predicted gene, 24457 |
4248 |
0.14 |
| chr15_78804542_78804702 | 0.13 |
Card10 |
caspase recruitment domain family, member 10 |
1580 |
0.24 |
| chr1_140018825_140019186 | 0.13 |
Gm26048 |
predicted gene, 26048 |
15180 |
0.15 |
| chr18_55506021_55506172 | 0.13 |
Gm37337 |
predicted gene, 37337 |
11018 |
0.3 |
| chr8_53848667_53848818 | 0.13 |
Gm19921 |
predicted gene, 19921 |
62997 |
0.15 |
| chr4_71808539_71808700 | 0.13 |
Gm11232 |
predicted gene 11232 |
50654 |
0.15 |
| chr12_37906736_37906912 | 0.13 |
Dgkb |
diacylglycerol kinase, beta |
26108 |
0.21 |
| chr14_57375309_57375470 | 0.13 |
Gm29717 |
predicted gene, 29717 |
9861 |
0.14 |
| chr11_60896053_60896204 | 0.13 |
Tmem11 |
transmembrane protein 11 |
16856 |
0.11 |
| chr1_24612181_24612351 | 0.13 |
Gm28439 |
predicted gene 28439 |
144 |
0.72 |
| chr19_39648534_39649207 | 0.13 |
Cyp2c67 |
cytochrome P450, family 2, subfamily c, polypeptide 67 |
181 |
0.97 |
| chr4_121002416_121002588 | 0.13 |
Smap2 |
small ArfGAP 2 |
14745 |
0.12 |
| chr14_70393692_70393872 | 0.13 |
Gm22725 |
predicted gene, 22725 |
28498 |
0.08 |
| chr13_28519541_28519699 | 0.13 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
1488 |
0.46 |
| chr4_9424353_9424518 | 0.13 |
Gm11817 |
predicted gene 11817 |
22764 |
0.21 |
| chr13_31440719_31440893 | 0.13 |
Gm23351 |
predicted gene, 23351 |
13551 |
0.17 |
| chr8_84068457_84068623 | 0.13 |
C330011M18Rik |
RIKEN cDNA C330011M18 gene |
1253 |
0.19 |
| chr10_22402809_22402960 | 0.13 |
Gm10825 |
predicted gene 10825 |
1 |
0.97 |
| chr10_118705843_118706313 | 0.13 |
Gm33337 |
predicted gene, 33337 |
9648 |
0.19 |
| chr8_8819298_8819591 | 0.13 |
Gm44622 |
predicted gene 44622 |
83474 |
0.06 |
| chrX_100470864_100471043 | 0.13 |
Igbp1 |
immunoglobulin (CD79A) binding protein 1 |
23338 |
0.13 |
| chrX_99292874_99293036 | 0.13 |
Gm14809 |
predicted gene 14809 |
125940 |
0.05 |
| chr5_117995379_117995641 | 0.13 |
Fbxo21 |
F-box protein 21 |
16186 |
0.13 |
| chr5_115866411_115866613 | 0.13 |
Gm42473 |
predicted gene 42473 |
3632 |
0.2 |
| chr18_11851878_11852035 | 0.13 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
7656 |
0.17 |
| chr12_103681317_103681508 | 0.13 |
Serpina16 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
5273 |
0.11 |
| chr6_85254139_85254316 | 0.13 |
Sfxn5 |
sideroflexin 5 |
10706 |
0.16 |
| chr8_36820142_36820568 | 0.12 |
Dlc1 |
deleted in liver cancer 1 |
87301 |
0.09 |
| chr19_5028205_5028356 | 0.12 |
Slc29a2 |
solute carrier family 29 (nucleoside transporters), member 2 |
2103 |
0.12 |
| chr9_52082020_52082183 | 0.12 |
Gm27686 |
predicted gene, 27686 |
21678 |
0.16 |
| chr10_68533323_68533612 | 0.12 |
Gm32872 |
predicted gene, 32872 |
3373 |
0.28 |
| chr18_6411106_6411281 | 0.12 |
Epc1 |
enhancer of polycomb homolog 1 |
30701 |
0.14 |
| chr6_34610404_34610696 | 0.12 |
Cald1 |
caldesmon 1 |
11930 |
0.19 |
| chr5_60042871_60043030 | 0.12 |
Gm43393 |
predicted gene 43393 |
17067 |
0.17 |
| chr16_45871401_45871600 | 0.12 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
27122 |
0.17 |
| chr10_125785978_125786129 | 0.12 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
180115 |
0.03 |
| chr2_75202592_75202875 | 0.12 |
Gm13653 |
predicted gene 13653 |
10456 |
0.15 |
| chr4_115496362_115496756 | 0.12 |
Cyp4a14 |
cytochrome P450, family 4, subfamily a, polypeptide 14 |
417 |
0.75 |
| chr5_8055710_8055890 | 0.12 |
Gm15730 |
predicted gene 15730 |
661 |
0.43 |
| chr11_31838871_31839199 | 0.12 |
Gm12107 |
predicted gene 12107 |
6375 |
0.2 |
| chr19_30147145_30147358 | 0.12 |
Gldc |
glycine decarboxylase |
2020 |
0.33 |
| chr1_139660727_139661253 | 0.12 |
Cfhr3 |
complement factor H-related 3 |
91 |
0.97 |
| chr11_100819438_100819589 | 0.12 |
Stat5b |
signal transducer and activator of transcription 5B |
3015 |
0.18 |
| chr15_6248496_6248831 | 0.12 |
Dab2 |
disabled 2, mitogen-responsive phosphoprotein |
51125 |
0.13 |
| chr2_131180305_131180466 | 0.12 |
Cenpb |
centromere protein B |
318 |
0.78 |
| chr4_118233934_118234146 | 0.12 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
1976 |
0.29 |
| chr11_62549816_62550114 | 0.12 |
Gm12279 |
predicted gene 12279 |
1076 |
0.25 |
| chr13_58058115_58058280 | 0.12 |
Mir874 |
microRNA 874 |
34997 |
0.13 |
| chr2_69140202_69140520 | 0.12 |
Nostrin |
nitric oxide synthase trafficker |
4561 |
0.23 |
| chr2_158783086_158783239 | 0.12 |
Fam83d |
family with sequence similarity 83, member D |
1754 |
0.34 |
| chr19_41523184_41523354 | 0.12 |
Lcor |
ligand dependent nuclear receptor corepressor |
26370 |
0.18 |
| chr12_5204482_5204649 | 0.12 |
Gm48532 |
predicted gene, 48532 |
31399 |
0.2 |
| chr4_43587824_43588150 | 0.12 |
Gm12472 |
predicted gene 12472 |
85 |
0.92 |
| chr19_4793945_4794103 | 0.12 |
Rbm4 |
RNA binding motif protein 4 |
123 |
0.91 |
| chr19_55251681_55251990 | 0.12 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
103 |
0.96 |
| chr6_51546167_51546340 | 0.12 |
Snx10 |
sorting nexin 10 |
1687 |
0.41 |
| chr6_142564803_142565069 | 0.12 |
Kcnj8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
6435 |
0.21 |
| chr19_39687799_39687975 | 0.12 |
Cyp2c67 |
cytochrome P450, family 2, subfamily c, polypeptide 67 |
38836 |
0.17 |
| chr11_68970449_68970600 | 0.12 |
Slc25a35 |
solute carrier family 25, member 35 |
2376 |
0.12 |
| chr11_83881252_83881469 | 0.12 |
Hnf1b |
HNF1 homeobox B |
28400 |
0.12 |
| chr4_135494462_135494778 | 0.12 |
Nipal3 |
NIPA-like domain containing 3 |
3 |
0.68 |
| chr13_38593226_38593439 | 0.12 |
Gm40922 |
predicted gene, 40922 |
38467 |
0.09 |
| chr15_61562347_61562502 | 0.12 |
Gm49498 |
predicted gene, 49498 |
18241 |
0.28 |
| chr3_135675157_135675328 | 0.12 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
5903 |
0.21 |
| chr16_24364185_24364362 | 0.12 |
Gm41434 |
predicted gene, 41434 |
5959 |
0.19 |
| chr4_40852306_40852662 | 0.12 |
B4galt1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
1521 |
0.21 |
| chr11_16701610_16702161 | 0.12 |
Gm25698 |
predicted gene, 25698 |
30826 |
0.15 |
| chr12_17738535_17738686 | 0.11 |
Hpcal1 |
hippocalcin-like 1 |
10384 |
0.23 |
| chr11_31822125_31822296 | 0.11 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
2263 |
0.31 |
| chr10_68268511_68268720 | 0.11 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
10106 |
0.23 |
| chr6_28891682_28891856 | 0.11 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
11726 |
0.21 |
| chr18_39855902_39856245 | 0.11 |
Gm41708 |
predicted gene, 41708 |
73301 |
0.1 |
| chr2_33178739_33178902 | 0.11 |
Gm13528 |
predicted gene 13528 |
918 |
0.51 |
| chr6_86127644_86127819 | 0.11 |
Gm19596 |
predicted gene, 19596 |
19421 |
0.13 |
| chr15_54947760_54948020 | 0.11 |
Gm26684 |
predicted gene, 26684 |
4180 |
0.19 |
| chr2_178711802_178711999 | 0.11 |
Cdh26 |
cadherin-like 26 |
251270 |
0.02 |
| chr10_53753298_53753451 | 0.11 |
Fam184a |
family with sequence similarity 184, member A |
2251 |
0.35 |
| chr7_130015298_130015740 | 0.11 |
Gm23847 |
predicted gene, 23847 |
19575 |
0.24 |
| chr6_6457346_6457728 | 0.11 |
Gm20685 |
predicted gene 20685 |
25485 |
0.17 |
| chr5_77098354_77098512 | 0.11 |
Hopx |
HOP homeobox |
3156 |
0.17 |
| chr5_115268690_115268875 | 0.11 |
Rnf10 |
ring finger protein 10 |
3519 |
0.1 |
| chr14_20177622_20177781 | 0.11 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
4108 |
0.16 |
| chr9_112743581_112744166 | 0.11 |
Gm24957 |
predicted gene, 24957 |
221344 |
0.02 |
| chr14_79511535_79511686 | 0.11 |
Elf1 |
E74-like factor 1 |
4064 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.2 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.2 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0018660 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |