Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
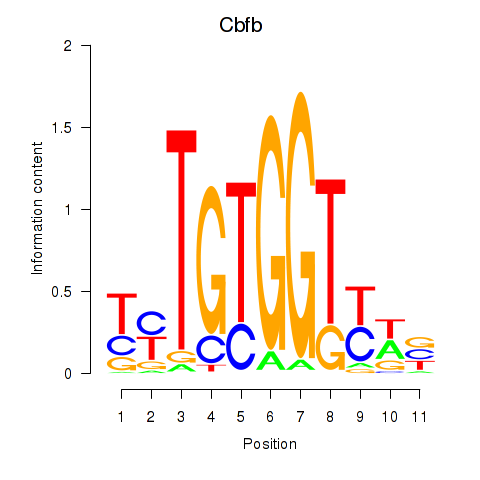
Results for Cbfb
Z-value: 0.56
Transcription factors associated with Cbfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cbfb
|
ENSMUSG00000031885.7 | core binding factor beta |
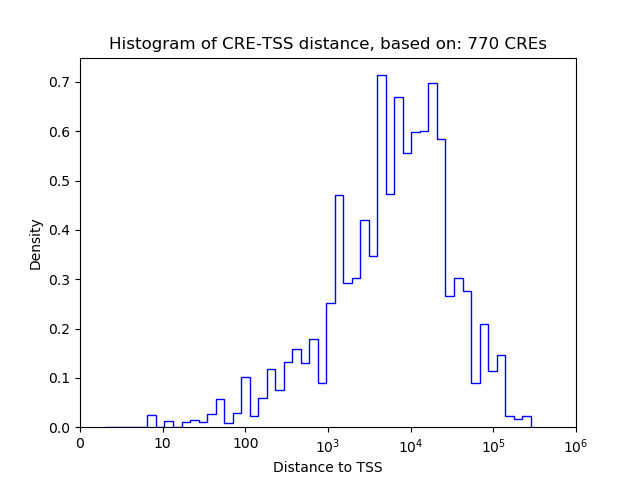
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_105171632_105171820 | Cbfb | 162 | 0.843457 | -0.67 | 1.5e-01 | Click! |
| chr8_105170139_105170326 | Cbfb | 442 | 0.664693 | -0.61 | 2.0e-01 | Click! |
| chr8_105171853_105172004 | Cbfb | 364 | 0.565879 | -0.57 | 2.4e-01 | Click! |
| chr8_105170450_105170615 | Cbfb | 142 | 0.904822 | 0.45 | 3.7e-01 | Click! |
| chr8_105169758_105169909 | Cbfb | 841 | 0.401169 | -0.16 | 7.6e-01 | Click! |
Activity of the Cbfb motif across conditions
Conditions sorted by the z-value of the Cbfb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
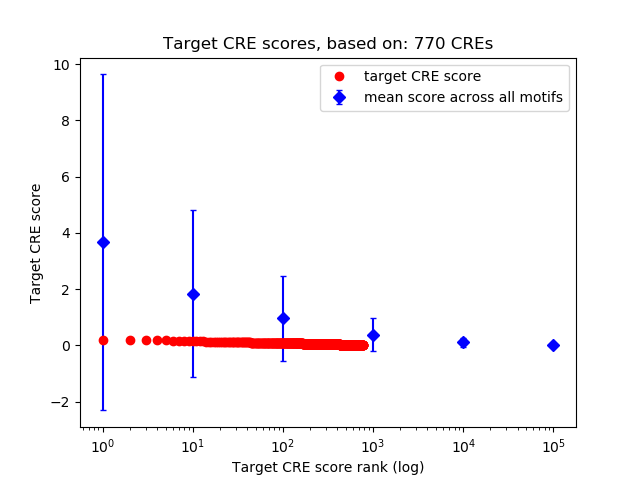
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_59893097_59893271 | 0.20 |
Gm19866 |
predicted gene, 19866 |
13935 |
0.13 |
| chr6_112939448_112939605 | 0.20 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
7228 |
0.15 |
| chr1_45826928_45827079 | 0.19 |
Wdr75 |
WD repeat domain 75 |
4445 |
0.16 |
| chr5_66114850_66115151 | 0.19 |
Rbm47 |
RNA binding motif protein 47 |
16809 |
0.11 |
| chr11_80453320_80453471 | 0.18 |
Psmd11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
2973 |
0.25 |
| chr6_55138486_55138637 | 0.17 |
Crhr2 |
corticotropin releasing hormone receptor 2 |
5545 |
0.18 |
| chr8_111739071_111739223 | 0.17 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
4662 |
0.21 |
| chr7_115686790_115686973 | 0.16 |
Sox6 |
SRY (sex determining region Y)-box 6 |
24516 |
0.27 |
| chr8_77045181_77045332 | 0.15 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
82757 |
0.09 |
| chr6_127265923_127266082 | 0.15 |
Gm43635 |
predicted gene 43635 |
11358 |
0.12 |
| chr11_4984826_4985069 | 0.14 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
1877 |
0.26 |
| chr9_40777068_40777228 | 0.14 |
Clmp |
CXADR-like membrane protein |
3054 |
0.12 |
| chr17_42900854_42901005 | 0.14 |
Cd2ap |
CD2-associated protein |
24264 |
0.24 |
| chr5_114378364_114378515 | 0.13 |
Kctd10 |
potassium channel tetramerisation domain containing 10 |
2044 |
0.21 |
| chr1_184605964_184606231 | 0.12 |
Rpl21-ps1 |
ribosomal protein 21, pseudogene 1 |
11151 |
0.15 |
| chr9_94993733_94993884 | 0.12 |
Gm47168 |
predicted gene, 47168 |
1124 |
0.65 |
| chr5_144126093_144126244 | 0.12 |
Gm26925 |
predicted gene, 26925 |
14396 |
0.14 |
| chr2_68884019_68884170 | 0.12 |
Gm37159 |
predicted gene, 37159 |
7272 |
0.15 |
| chr6_128646065_128646273 | 0.12 |
Gm5884 |
predicted pseudogene 5884 |
1256 |
0.23 |
| chr16_31419378_31420071 | 0.12 |
Bdh1 |
3-hydroxybutyrate dehydrogenase, type 1 |
2556 |
0.19 |
| chr1_119173195_119173378 | 0.12 |
Gm8321 |
predicted gene 8321 |
15700 |
0.22 |
| chr4_149282398_149282549 | 0.11 |
Kif1b |
kinesin family member 1B |
25033 |
0.13 |
| chr10_5592140_5592291 | 0.11 |
Myct1 |
myc target 1 |
1560 |
0.45 |
| chr12_105686564_105686722 | 0.11 |
Gskip |
GSK3B interacting protein |
1242 |
0.32 |
| chr1_74400693_74400993 | 0.11 |
Mir26b |
microRNA 26b |
6533 |
0.11 |
| chr18_39251064_39251253 | 0.11 |
Arhgap26 |
Rho GTPase activating protein 26 |
23790 |
0.21 |
| chr2_75611169_75611330 | 0.11 |
Gm13655 |
predicted gene 13655 |
22133 |
0.15 |
| chr7_122665413_122665987 | 0.11 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
4792 |
0.21 |
| chr1_138967552_138967703 | 0.11 |
Dennd1b |
DENN/MADD domain containing 1B |
93 |
0.91 |
| chr6_141866172_141866335 | 0.10 |
Gm30784 |
predicted gene, 30784 |
105 |
0.97 |
| chr19_26748788_26748982 | 0.10 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
353 |
0.9 |
| chr1_31019080_31019348 | 0.10 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
19310 |
0.13 |
| chr11_75459071_75459222 | 0.10 |
Mir22hg |
Mir22 host gene (non-protein coding) |
2393 |
0.12 |
| chr13_64313921_64314144 | 0.10 |
Prxl2c |
peroxiredoxin like 2C |
1322 |
0.27 |
| chr10_67185588_67186325 | 0.10 |
Jmjd1c |
jumonji domain containing 1C |
203 |
0.95 |
| chr2_104817028_104817755 | 0.10 |
Qser1 |
glutamine and serine rich 1 |
631 |
0.64 |
| chr6_128503302_128503638 | 0.10 |
Pzp |
PZP, alpha-2-macroglobulin like |
8247 |
0.09 |
| chr3_57431826_57431986 | 0.10 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
6592 |
0.23 |
| chr15_27528988_27529148 | 0.10 |
B230362B09Rik |
RIKEN cDNA B230362B09 gene |
25156 |
0.14 |
| chr9_116847373_116847540 | 0.10 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
24637 |
0.27 |
| chr2_168314984_168315140 | 0.10 |
Gm14262 |
predicted gene 14262 |
19941 |
0.13 |
| chr4_118033071_118033392 | 0.10 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
1349 |
0.44 |
| chr9_110362456_110362638 | 0.10 |
Scap |
SREBF chaperone |
930 |
0.43 |
| chr3_107223260_107223437 | 0.10 |
Cym |
chymosin |
1616 |
0.29 |
| chr11_38681374_38681545 | 0.10 |
Gm23520 |
predicted gene, 23520 |
116818 |
0.07 |
| chr12_100528331_100528561 | 0.10 |
Ttc7b |
tetratricopeptide repeat domain 7B |
7620 |
0.17 |
| chr10_59901049_59901200 | 0.10 |
Dnajb12 |
DnaJ heat shock protein family (Hsp40) member B12 |
21498 |
0.14 |
| chr19_39284209_39284379 | 0.10 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
2780 |
0.3 |
| chr19_34819627_34820189 | 0.10 |
Mir107 |
microRNA 107 |
865 |
0.51 |
| chr7_81326206_81326373 | 0.10 |
Rps17 |
ribosomal protein S17 |
18242 |
0.14 |
| chr13_95894500_95894710 | 0.10 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
2122 |
0.3 |
| chr14_66199873_66200054 | 0.09 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
13589 |
0.19 |
| chr12_110757292_110757448 | 0.09 |
Wdr20 |
WD repeat domain 20 |
19161 |
0.13 |
| chr19_31896057_31896235 | 0.09 |
Gm19241 |
predicted gene, 19241 |
17498 |
0.2 |
| chr4_83167479_83167646 | 0.09 |
Gm11185 |
predicted gene 11185 |
26426 |
0.17 |
| chr11_61389045_61389196 | 0.09 |
Slc47a1 |
solute carrier family 47, member 1 |
10775 |
0.13 |
| chr17_80921778_80921949 | 0.09 |
Gm9386 |
predicted pseudogene 9386 |
16851 |
0.18 |
| chr2_12011493_12011801 | 0.09 |
Gm13310 |
predicted gene 13310 |
72345 |
0.1 |
| chr8_31980849_31981024 | 0.09 |
Nrg1 |
neuregulin 1 |
28170 |
0.23 |
| chr1_193102685_193102845 | 0.09 |
Utp25 |
UTP25 small subunit processome component |
11910 |
0.14 |
| chr18_54921904_54922070 | 0.09 |
Zfp608 |
zinc finger protein 608 |
26425 |
0.17 |
| chr1_118458813_118459291 | 0.09 |
Gm22710 |
predicted gene, 22710 |
213 |
0.44 |
| chr3_96528686_96528855 | 0.09 |
Hjv |
hemojuvelin BMP co-receptor |
3598 |
0.09 |
| chr10_125317485_125317647 | 0.09 |
Gm45604 |
predicted gene 45604 |
8654 |
0.23 |
| chr8_115761720_115761892 | 0.09 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
54012 |
0.15 |
| chr3_132565872_132566040 | 0.09 |
Gm25307 |
predicted gene, 25307 |
28291 |
0.18 |
| chr1_106309746_106309897 | 0.09 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
7417 |
0.25 |
| chr4_57916402_57916601 | 0.09 |
D630039A03Rik |
RIKEN cDNA D630039A03 gene |
204 |
0.94 |
| chr16_4405370_4405807 | 0.09 |
Adcy9 |
adenylate cyclase 9 |
13999 |
0.21 |
| chr4_105104623_105104776 | 0.09 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
5191 |
0.29 |
| chr1_140183261_140183634 | 0.09 |
Cfh |
complement component factor h |
36 |
0.98 |
| chr6_38866474_38866676 | 0.09 |
Tbxas1 |
thromboxane A synthase 1, platelet |
8829 |
0.21 |
| chr4_41493635_41493797 | 0.09 |
Myorg |
myogenesis regulating glycosidase (putative) |
8952 |
0.1 |
| chr1_93723491_93723642 | 0.09 |
Gm10550 |
predicted gene 10550 |
7538 |
0.13 |
| chr15_82671625_82671923 | 0.09 |
Cyp2d36-ps |
cytochrome P450, family 2, subfamily d, polypeptide 36, pseudogene |
11483 |
0.08 |
| chr11_32688599_32688774 | 0.09 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
45782 |
0.14 |
| chr6_15728953_15729104 | 0.09 |
Mdfic |
MyoD family inhibitor domain containing |
1229 |
0.59 |
| chr9_102410773_102410941 | 0.09 |
Gm22894 |
predicted gene, 22894 |
37739 |
0.13 |
| chr13_104650268_104650432 | 0.09 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
34581 |
0.22 |
| chr18_63463560_63463749 | 0.09 |
Gm23581 |
predicted gene, 23581 |
44515 |
0.11 |
| chr1_127269428_127269591 | 0.08 |
Gm37510 |
predicted gene, 37510 |
1015 |
0.5 |
| chr4_139208375_139208554 | 0.08 |
Capzb |
capping protein (actin filament) muscle Z-line, beta |
9197 |
0.15 |
| chr19_23024753_23025259 | 0.08 |
Gm50136 |
predicted gene, 50136 |
36448 |
0.17 |
| chr15_79741187_79741771 | 0.08 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
613 |
0.45 |
| chr12_25190485_25190636 | 0.08 |
Gm47705 |
predicted gene, 47705 |
8792 |
0.19 |
| chr19_29180439_29180590 | 0.08 |
Gm5518 |
predicted gene 5518 |
20396 |
0.15 |
| chr16_74514689_74514840 | 0.08 |
Gm49671 |
predicted gene, 49671 |
91498 |
0.08 |
| chr6_86189199_86189379 | 0.08 |
Tgfa |
transforming growth factor alpha |
5934 |
0.17 |
| chr4_115497333_115497797 | 0.08 |
Cyp4a14 |
cytochrome P450, family 4, subfamily a, polypeptide 14 |
1423 |
0.29 |
| chr15_59721710_59721861 | 0.08 |
Gm20150 |
predicted gene, 20150 |
49672 |
0.14 |
| chr8_48690628_48690947 | 0.08 |
Tenm3 |
teneurin transmembrane protein 3 |
16097 |
0.26 |
| chr11_87984674_87984835 | 0.08 |
Dynll2 |
dynein light chain LC8-type 2 |
742 |
0.57 |
| chr17_13402789_13403038 | 0.08 |
Gm8597 |
predicted gene 8597 |
1278 |
0.29 |
| chr13_44293994_44294145 | 0.08 |
Gm29676 |
predicted gene, 29676 |
13620 |
0.2 |
| chr10_69930149_69930514 | 0.08 |
Ank3 |
ankyrin 3, epithelial |
4198 |
0.35 |
| chr1_167247272_167247459 | 0.08 |
Uck2 |
uridine-cytidine kinase 2 |
4207 |
0.15 |
| chr10_80914031_80914448 | 0.08 |
Lmnb2 |
lamin B2 |
3612 |
0.11 |
| chr14_48714246_48714540 | 0.08 |
Gm49153 |
predicted gene, 49153 |
10209 |
0.1 |
| chr8_128481354_128481549 | 0.08 |
Nrp1 |
neuropilin 1 |
122054 |
0.05 |
| chr18_38631612_38631778 | 0.08 |
9630014M24Rik |
RIKEN cDNA 9630014M24 gene |
4408 |
0.19 |
| chr6_113721227_113721413 | 0.08 |
Ghrl |
ghrelin |
1440 |
0.19 |
| chr5_38480616_38480794 | 0.08 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
399 |
0.85 |
| chr2_163411853_163412159 | 0.08 |
Oser1 |
oxidative stress responsive serine rich 1 |
341 |
0.82 |
| chr11_97920651_97920808 | 0.08 |
Gm22461 |
predicted gene, 22461 |
21011 |
0.1 |
| chr9_90260660_90260839 | 0.08 |
Tbc1d2b |
TBC1 domain family, member 2B |
4822 |
0.19 |
| chr3_108134800_108134966 | 0.08 |
Gnai3 |
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
11263 |
0.09 |
| chr2_12905587_12905857 | 0.08 |
Pter |
phosphotriesterase related |
18319 |
0.22 |
| chr12_79642030_79642203 | 0.08 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
282617 |
0.01 |
| chr5_137628625_137628972 | 0.08 |
Fbxo24 |
F-box protein 24 |
158 |
0.55 |
| chr4_86664611_86664778 | 0.08 |
Plin2 |
perilipin 2 |
1031 |
0.52 |
| chr17_86203109_86203281 | 0.08 |
AC154542.1 |
TEC |
27620 |
0.18 |
| chr3_65533694_65533867 | 0.08 |
4931440P22Rik |
RIKEN cDNA 4931440P22 gene |
4401 |
0.15 |
| chr4_117981622_117981822 | 0.08 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
6686 |
0.15 |
| chr8_113202539_113202735 | 0.07 |
Gm10280 |
predicted gene 10280 |
135375 |
0.05 |
| chr10_118797429_118797609 | 0.07 |
Gm4065 |
predicted gene 4065 |
7740 |
0.17 |
| chr5_91274508_91274681 | 0.07 |
Gm19619 |
predicted gene, 19619 |
8827 |
0.24 |
| chr9_31020201_31020366 | 0.07 |
Gm29724 |
predicted gene, 29724 |
10220 |
0.17 |
| chr2_14130166_14130332 | 0.07 |
Stam |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
595 |
0.69 |
| chr11_84476404_84476555 | 0.07 |
Aatf |
apoptosis antagonizing transcription factor |
15071 |
0.21 |
| chr13_97239621_97239811 | 0.07 |
Enc1 |
ectodermal-neural cortex 1 |
1389 |
0.38 |
| chr17_33563776_33563966 | 0.07 |
Myo1f |
myosin IF |
8106 |
0.14 |
| chr2_65441659_65441885 | 0.07 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
53459 |
0.11 |
| chr3_95894699_95894897 | 0.07 |
Aph1a |
aph1 homolog A, gamma secretase subunit |
333 |
0.72 |
| chr8_83738255_83738433 | 0.07 |
Adgre5 |
adhesion G protein-coupled receptor E5 |
2825 |
0.16 |
| chr1_136608978_136609129 | 0.07 |
4933409D19Rik |
RIKEN cDNA 4933409D19 gene |
3585 |
0.16 |
| chr14_54433612_54433789 | 0.07 |
Mmp14 |
matrix metallopeptidase 14 (membrane-inserted) |
1264 |
0.24 |
| chr5_43904829_43905012 | 0.07 |
Gm16015 |
predicted gene 16015 |
4965 |
0.11 |
| chr13_100849368_100849536 | 0.07 |
Gm29502 |
predicted gene 29502 |
2019 |
0.24 |
| chr11_95461001_95461320 | 0.07 |
Gm11522 |
predicted gene 11522 |
1561 |
0.33 |
| chr12_70280483_70280649 | 0.07 |
Trim9 |
tripartite motif-containing 9 |
12518 |
0.16 |
| chr5_134974335_134974526 | 0.07 |
Cldn3 |
claudin 3 |
11784 |
0.07 |
| chr11_90371567_90371726 | 0.07 |
Hlf |
hepatic leukemia factor |
10289 |
0.26 |
| chr19_55660221_55660414 | 0.07 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
81503 |
0.11 |
| chr7_97211249_97211431 | 0.07 |
Usp35 |
ubiquitin specific peptidase 35 |
103685 |
0.06 |
| chr11_55471153_55471304 | 0.07 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1510 |
0.25 |
| chr18_74741418_74741569 | 0.07 |
Myo5b |
myosin VB |
7728 |
0.18 |
| chr5_148936597_148936854 | 0.07 |
Katnal1 |
katanin p60 subunit A-like 1 |
7405 |
0.1 |
| chr14_61294825_61294981 | 0.07 |
Gm19097 |
predicted gene, 19097 |
10115 |
0.12 |
| chr12_111435070_111435221 | 0.07 |
Gm10425 |
predicted gene 10425 |
4199 |
0.14 |
| chr2_122237988_122238172 | 0.07 |
Sord |
sorbitol dehydrogenase |
3331 |
0.14 |
| chr18_20552765_20552968 | 0.07 |
Tpi-rs10 |
triosephosphate isomerase related sequence 10 |
3502 |
0.21 |
| chr3_60516106_60516484 | 0.07 |
Mbnl1 |
muscleblind like splicing factor 1 |
11833 |
0.22 |
| chr9_70827748_70828187 | 0.07 |
Gm3436 |
predicted pseudogene 3436 |
24609 |
0.16 |
| chr19_31896808_31896966 | 0.07 |
Gm19241 |
predicted gene, 19241 |
16757 |
0.2 |
| chr12_75668168_75668511 | 0.07 |
Wdr89 |
WD repeat domain 89 |
1188 |
0.51 |
| chr18_31884190_31884397 | 0.07 |
Wdr33 |
WD repeat domain 33 |
2151 |
0.25 |
| chr18_34864822_34864973 | 0.07 |
Egr1 |
early growth response 1 |
3690 |
0.17 |
| chr4_155123910_155124068 | 0.07 |
Morn1 |
MORN repeat containing 1 |
13299 |
0.15 |
| chr7_27474827_27474987 | 0.07 |
Sertad3 |
SERTA domain containing 3 |
1139 |
0.28 |
| chr10_108619351_108620201 | 0.07 |
Syt1 |
synaptotagmin I |
17188 |
0.25 |
| chr2_26543305_26543481 | 0.07 |
Gm13358 |
predicted gene 13358 |
15232 |
0.09 |
| chr8_108887457_108887635 | 0.07 |
Gm38318 |
predicted gene, 38318 |
19984 |
0.2 |
| chr19_43949859_43950010 | 0.07 |
Dnmbp |
dynamin binding protein |
9743 |
0.13 |
| chr19_29667197_29667356 | 0.07 |
Ermp1 |
endoplasmic reticulum metallopeptidase 1 |
18861 |
0.17 |
| chr5_121659952_121660313 | 0.07 |
Acad10 |
acyl-Coenzyme A dehydrogenase family, member 10 |
347 |
0.52 |
| chr18_80275077_80275246 | 0.07 |
Slc66a2 |
solute carrier family 66 member 2 |
12321 |
0.11 |
| chr9_42998416_42998567 | 0.07 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
28169 |
0.15 |
| chr14_59485330_59485520 | 0.07 |
Cab39l |
calcium binding protein 39-like |
6609 |
0.21 |
| chr4_130774522_130774687 | 0.07 |
Gm23711 |
predicted gene, 23711 |
1137 |
0.34 |
| chr1_88587324_88587676 | 0.07 |
Gm19589 |
predicted gene, 19589 |
35983 |
0.13 |
| chr7_132660508_132660703 | 0.07 |
Gm15582 |
predicted gene 15582 |
721 |
0.66 |
| chr6_115658927_115659091 | 0.07 |
Raf1 |
v-raf-leukemia viral oncogene 1 |
16955 |
0.11 |
| chr7_35557659_35557843 | 0.07 |
Nudt19 |
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
1447 |
0.28 |
| chr10_95678111_95678450 | 0.07 |
Anapc15-ps |
anaphase promoting complex C subunit 15, pseudogene |
4829 |
0.14 |
| chr19_23072349_23072500 | 0.06 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
3429 |
0.24 |
| chr4_94725954_94726107 | 0.06 |
Tek |
TEK receptor tyrosine kinase |
13259 |
0.19 |
| chr7_16254402_16254580 | 0.06 |
C5ar1 |
complement component 5a receptor 1 |
4847 |
0.11 |
| chr18_30750845_30751026 | 0.06 |
Gm41780 |
predicted gene, 41780 |
142045 |
0.05 |
| chr6_71193984_71194261 | 0.06 |
Fabp1 |
fatty acid binding protein 1, liver |
5705 |
0.14 |
| chr2_50984081_50984271 | 0.06 |
Gm13498 |
predicted gene 13498 |
74492 |
0.12 |
| chr15_27782797_27782987 | 0.06 |
Trio |
triple functional domain (PTPRF interacting) |
5746 |
0.25 |
| chr4_97873061_97873239 | 0.06 |
Nfia |
nuclear factor I/A |
6969 |
0.29 |
| chr3_92440282_92440451 | 0.06 |
Sprr1b |
small proline-rich protein 1B |
1577 |
0.18 |
| chr8_86538609_86538876 | 0.06 |
Abcc12 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
3995 |
0.27 |
| chr19_56774484_56774710 | 0.06 |
Gm6990 |
predicted pseudogene 6990 |
19251 |
0.14 |
| chr2_72222793_72223023 | 0.06 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
9394 |
0.17 |
| chr16_28886318_28886476 | 0.06 |
Mb21d2 |
Mab-21 domain containing 2 |
43276 |
0.18 |
| chr1_170516583_170516766 | 0.06 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
40436 |
0.16 |
| chr9_50877506_50877697 | 0.06 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
484 |
0.77 |
| chr16_91869715_91869882 | 0.06 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
7755 |
0.16 |
| chr1_13267456_13267729 | 0.06 |
Gm27881 |
predicted gene, 27881 |
5394 |
0.14 |
| chr7_19266055_19266214 | 0.06 |
Vasp |
vasodilator-stimulated phosphoprotein |
412 |
0.65 |
| chr8_115856888_115857171 | 0.06 |
Gm45733 |
predicted gene 45733 |
115042 |
0.06 |
| chr14_62329558_62329856 | 0.06 |
Rnaseh2b |
ribonuclease H2, subunit B |
2361 |
0.32 |
| chr9_104331483_104331659 | 0.06 |
Acpp |
acid phosphatase, prostate |
6111 |
0.17 |
| chr6_115901113_115901499 | 0.06 |
Ift122 |
intraflagellar transport 122 |
1321 |
0.32 |
| chr11_111944397_111944560 | 0.06 |
Gm11679 |
predicted gene 11679 |
99100 |
0.08 |
| chr19_42144194_42144345 | 0.06 |
Marveld1 |
MARVEL (membrane-associating) domain containing 1 |
3131 |
0.15 |
| chr8_110939430_110939581 | 0.06 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
1399 |
0.28 |
| chr6_54762292_54762450 | 0.06 |
Znrf2 |
zinc and ring finger 2 |
54545 |
0.11 |
| chr16_22886551_22886735 | 0.06 |
Ahsg |
alpha-2-HS-glycoprotein |
4634 |
0.13 |
| chr8_35415233_35415420 | 0.06 |
Gm45301 |
predicted gene 45301 |
5820 |
0.18 |
| chr13_55369651_55369802 | 0.06 |
Rgs14 |
regulator of G-protein signaling 14 |
6 |
0.95 |
| chr5_3364806_3364983 | 0.06 |
Cdk6 |
cyclin-dependent kinase 6 |
20582 |
0.19 |
| chr1_38242754_38242912 | 0.06 |
Gm16150 |
predicted gene 16150 |
3133 |
0.27 |
| chr13_115102869_115103020 | 0.06 |
Itga1 |
integrin alpha 1 |
980 |
0.34 |
| chr17_43275309_43275499 | 0.06 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
5075 |
0.29 |
| chr9_112735889_112736061 | 0.06 |
Gm24957 |
predicted gene, 24957 |
213446 |
0.02 |
| chr19_55808091_55808481 | 0.06 |
Ppnr |
per-pentamer repeat gene |
33386 |
0.21 |
| chr4_47294478_47294667 | 0.06 |
Col15a1 |
collagen, type XV, alpha 1 |
6285 |
0.24 |
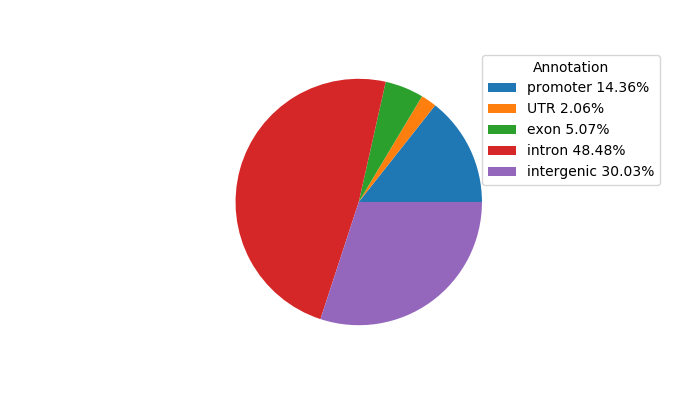
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |