Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
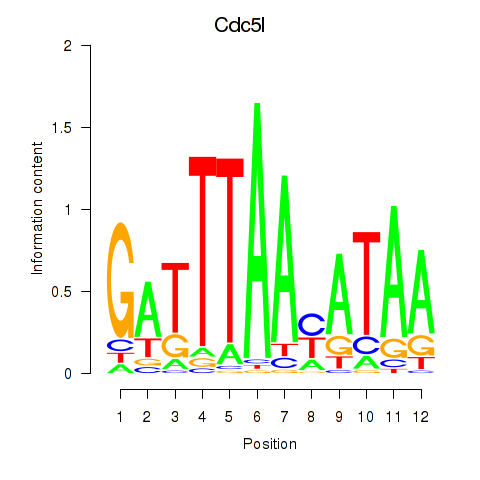
Results for Cdc5l
Z-value: 1.96
Transcription factors associated with Cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cdc5l
|
ENSMUSG00000023932.8 | cell division cycle 5-like (S. pombe) |
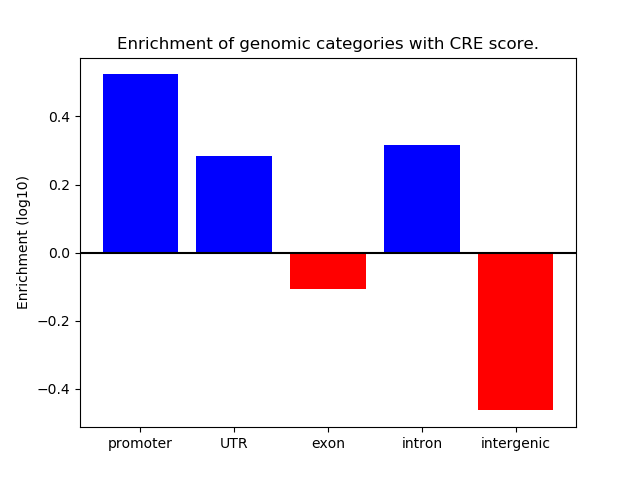
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_45432981_45433132 | Cdc5l | 485 | 0.473916 | -0.81 | 5.0e-02 | Click! |
| chr17_45322913_45323086 | Cdc5l | 92685 | 0.062487 | -0.76 | 7.7e-02 | Click! |
| chr17_45402203_45402541 | Cdc5l | 13312 | 0.164763 | 0.53 | 2.8e-01 | Click! |
| chr17_45402856_45403089 | Cdc5l | 12712 | 0.165237 | 0.52 | 2.9e-01 | Click! |
| chr17_45433278_45433437 | Cdc5l | 184 | 0.565485 | -0.51 | 3.0e-01 | Click! |
Activity of the Cdc5l motif across conditions
Conditions sorted by the z-value of the Cdc5l motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
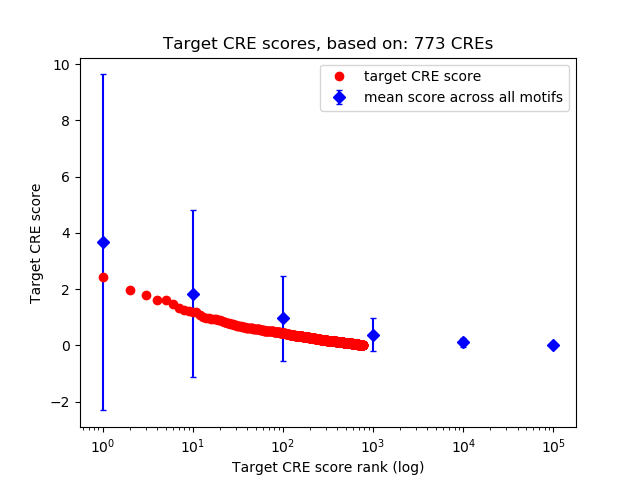
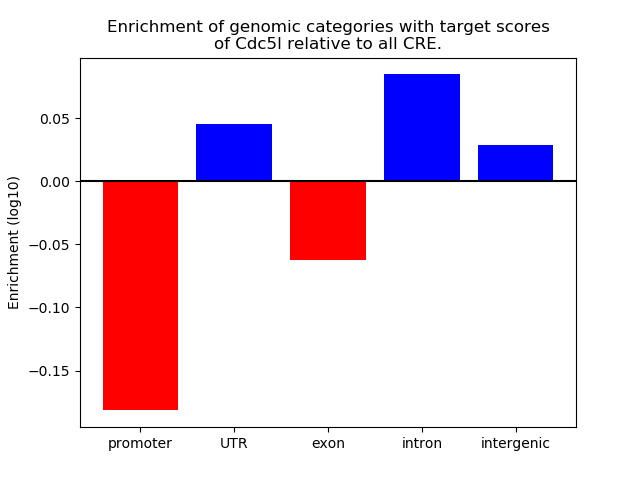
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_96674138_96674322 | 2.44 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
3294 |
0.19 |
| chr8_40878264_40878415 | 1.98 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
3391 |
0.2 |
| chr9_19622407_19622600 | 1.79 |
Zfp317 |
zinc finger protein 317 |
199 |
0.91 |
| chr2_65167070_65167237 | 1.60 |
Cobll1 |
Cobl-like 1 |
61294 |
0.12 |
| chr19_36263973_36264136 | 1.60 |
Gm32027 |
predicted gene, 32027 |
383 |
0.85 |
| chr7_73542824_73542975 | 1.46 |
Chd2 |
chromodomain helicase DNA binding protein 2 |
1069 |
0.35 |
| chr11_16803107_16803282 | 1.33 |
Egfros |
epidermal growth factor receptor, opposite strand |
27508 |
0.17 |
| chr6_119883269_119883450 | 1.27 |
Gm15532 |
predicted gene 15532 |
9561 |
0.15 |
| chr1_172159886_172160228 | 1.23 |
Dcaf8 |
DDB1 and CUL4 associated factor 8 |
11109 |
0.1 |
| chr2_52575707_52576080 | 1.20 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
17326 |
0.18 |
| chr19_40153581_40153732 | 1.19 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33630 |
0.13 |
| chr10_52420605_52420773 | 1.07 |
Nus1 |
NUS1 dehydrodolichyl diphosphate synthase subunit |
3142 |
0.15 |
| chr16_72808282_72808697 | 1.02 |
Robo1 |
roundabout guidance receptor 1 |
145285 |
0.05 |
| chr15_6218493_6218848 | 0.97 |
Gm23139 |
predicted gene, 23139 |
67322 |
0.11 |
| chr10_42723053_42723245 | 0.96 |
Gm15200 |
predicted gene 15200 |
24631 |
0.14 |
| chr17_50378350_50378501 | 0.94 |
Gm49906 |
predicted gene, 49906 |
39173 |
0.17 |
| chr17_24209788_24209956 | 0.94 |
Ntn3 |
netrin 3 |
341 |
0.67 |
| chr6_144070883_144071211 | 0.92 |
Sox5os1 |
SRY (sex determining region Y)-box 5, opposite strand 1 |
49139 |
0.17 |
| chr5_102010547_102010959 | 0.91 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29195 |
0.16 |
| chr3_60034453_60034756 | 0.89 |
Aadac |
arylacetamide deacetylase |
2728 |
0.23 |
| chr4_4131153_4131310 | 0.86 |
A830012C17Rik |
RIKEN cDNA A830012C17 gene |
7090 |
0.2 |
| chr14_122602774_122602990 | 0.85 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
37 |
0.98 |
| chr7_132967481_132967632 | 0.84 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
13303 |
0.14 |
| chr9_116136291_116136469 | 0.81 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38885 |
0.15 |
| chr15_42296635_42296795 | 0.78 |
Gm49452 |
predicted gene, 49452 |
23631 |
0.17 |
| chr8_48663447_48663609 | 0.76 |
Tenm3 |
teneurin transmembrane protein 3 |
11156 |
0.25 |
| chr9_83726172_83726335 | 0.75 |
Gm2109 |
predicted gene 2109 |
34778 |
0.15 |
| chr4_53099364_53099515 | 0.75 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
58513 |
0.11 |
| chr13_63113927_63114104 | 0.74 |
Aopep |
aminopeptidase O |
6493 |
0.16 |
| chr1_67168769_67168980 | 0.73 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
45848 |
0.15 |
| chr1_13374931_13375133 | 0.70 |
Ncoa2 |
nuclear receptor coactivator 2 |
949 |
0.34 |
| chr17_43276058_43276209 | 0.68 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
5804 |
0.29 |
| chr6_149149841_149149992 | 0.68 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
8270 |
0.13 |
| chr13_21236545_21236716 | 0.67 |
Trim27 |
tripartite motif-containing 27 |
46769 |
0.08 |
| chr2_58782122_58782273 | 0.66 |
Upp2 |
uridine phosphorylase 2 |
16872 |
0.2 |
| chr2_72752692_72753055 | 0.65 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
2020 |
0.38 |
| chr9_106235817_106235968 | 0.65 |
Alas1 |
aminolevulinic acid synthase 1 |
1192 |
0.3 |
| chr3_77832077_77832234 | 0.64 |
Gm37576 |
predicted gene, 37576 |
40998 |
0.19 |
| chr5_147320087_147320238 | 0.63 |
Urad |
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
2278 |
0.17 |
| chr9_107305345_107305527 | 0.63 |
Gm17041 |
predicted gene 17041 |
3598 |
0.12 |
| chr15_53146756_53146975 | 0.62 |
Ext1 |
exostosin glycosyltransferase 1 |
38997 |
0.23 |
| chr18_66001524_66001695 | 0.62 |
Lman1 |
lectin, mannose-binding, 1 |
1009 |
0.44 |
| chr13_114974629_114975014 | 0.62 |
Gm47776 |
predicted gene, 47776 |
39753 |
0.15 |
| chr8_76152735_76152886 | 0.61 |
Gm45742 |
predicted gene 45742 |
35783 |
0.19 |
| chr4_61807807_61807958 | 0.61 |
Mup-ps16 |
major urinary protein, pseudogene 16 |
10400 |
0.12 |
| chr13_19395072_19395660 | 0.60 |
Gm42683 |
predicted gene 42683 |
87 |
0.82 |
| chr1_45922232_45922396 | 0.58 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
995 |
0.45 |
| chr11_118201294_118201477 | 0.58 |
Gm11737 |
predicted gene 11737 |
3794 |
0.18 |
| chr8_69396075_69396226 | 0.58 |
Gm10033 |
predicted gene 10033 |
611 |
0.64 |
| chr1_67192772_67192960 | 0.58 |
Gm15668 |
predicted gene 15668 |
56334 |
0.12 |
| chr8_22893059_22893230 | 0.58 |
Gm45555 |
predicted gene 45555 |
19555 |
0.14 |
| chrX_101952539_101952717 | 0.57 |
Nhsl2 |
NHS-like 2 |
9169 |
0.17 |
| chr1_67213641_67214357 | 0.57 |
Gm15668 |
predicted gene 15668 |
35201 |
0.17 |
| chr3_52011351_52011543 | 0.57 |
Gm37465 |
predicted gene, 37465 |
7422 |
0.13 |
| chr2_144550194_144550414 | 0.56 |
Rbbp9 |
retinoblastoma binding protein 9, serine hydrolase |
486 |
0.71 |
| chr19_36781247_36781436 | 0.56 |
Gm50112 |
predicted gene, 50112 |
37307 |
0.14 |
| chr2_160797467_160797706 | 0.56 |
Plcg1 |
phospholipase C, gamma 1 |
37468 |
0.11 |
| chr6_138156558_138156709 | 0.55 |
Mgst1 |
microsomal glutathione S-transferase 1 |
13779 |
0.27 |
| chr9_74791015_74791321 | 0.55 |
Gm22315 |
predicted gene, 22315 |
9098 |
0.19 |
| chr10_40511211_40511581 | 0.55 |
Gm18671 |
predicted gene, 18671 |
37496 |
0.14 |
| chr14_59443633_59443793 | 0.55 |
Cab39l |
calcium binding protein 39-like |
2732 |
0.21 |
| chr6_26490665_26490857 | 0.52 |
Gm24691 |
predicted gene, 24691 |
30331 |
0.26 |
| chr10_86690190_86690443 | 0.52 |
Gm15344 |
predicted gene 15344 |
2779 |
0.1 |
| chr10_107325957_107326150 | 0.51 |
Lin7a |
lin-7 homolog A (C. elegans) |
53659 |
0.14 |
| chr11_79884650_79884847 | 0.51 |
Gm24454 |
predicted gene, 24454 |
8967 |
0.14 |
| chr8_93073771_93073922 | 0.51 |
Ces1b |
carboxylesterase 1B |
6171 |
0.16 |
| chr18_9732141_9732318 | 0.51 |
Gm17430 |
predicted gene, 17430 |
5559 |
0.13 |
| chr18_59092890_59093288 | 0.51 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
30578 |
0.22 |
| chr18_12760468_12760619 | 0.50 |
Osbpl1a |
oxysterol binding protein-like 1A |
6594 |
0.15 |
| chr14_62921769_62921931 | 0.50 |
Gm21021 |
predicted gene, 21021 |
51940 |
0.08 |
| chr2_73278335_73278505 | 0.50 |
Sp9 |
trans-acting transcription factor 9 |
6454 |
0.16 |
| chr6_119681087_119681251 | 0.50 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
59064 |
0.13 |
| chr2_84648708_84648914 | 0.50 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
274 |
0.82 |
| chr19_31896510_31896684 | 0.50 |
Gm19241 |
predicted gene, 19241 |
17047 |
0.2 |
| chr14_47295909_47296231 | 0.50 |
Mapk1ip1l |
mitogen-activated protein kinase 1 interacting protein 1-like |
2196 |
0.13 |
| chr2_126934579_126934730 | 0.50 |
Sppl2a |
signal peptide peptidase like 2A |
1419 |
0.4 |
| chr1_67153777_67153928 | 0.50 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
30826 |
0.19 |
| chr14_11125061_11125216 | 0.49 |
Fhit |
fragile histidine triad gene |
41 |
0.98 |
| chr9_35060528_35060679 | 0.49 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
4809 |
0.2 |
| chr12_32936719_32936876 | 0.49 |
Gm47961 |
predicted gene, 47961 |
5065 |
0.13 |
| chr2_167841674_167841862 | 0.48 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
8122 |
0.16 |
| chr15_10177985_10178196 | 0.48 |
Prlr |
prolactin receptor |
309 |
0.94 |
| chr13_37831757_37832025 | 0.48 |
Rreb1 |
ras responsive element binding protein 1 |
4498 |
0.22 |
| chr15_36595891_36596298 | 0.48 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
1332 |
0.34 |
| chr4_53222171_53222454 | 0.48 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
4455 |
0.21 |
| chr4_6268102_6268325 | 0.48 |
Gm11798 |
predicted gene 11798 |
7248 |
0.2 |
| chr10_121501382_121501572 | 0.47 |
Gm46204 |
predicted gene, 46204 |
3874 |
0.13 |
| chr10_85957276_85957442 | 0.47 |
Rtcb |
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
464 |
0.7 |
| chr3_63962435_63962754 | 0.47 |
Gm26850 |
predicted gene, 26850 |
1039 |
0.38 |
| chr2_18435403_18435565 | 0.46 |
Dnajc1 |
DnaJ heat shock protein family (Hsp40) member C1 |
42654 |
0.16 |
| chr1_67207008_67207510 | 0.46 |
Gm15668 |
predicted gene 15668 |
41941 |
0.15 |
| chr11_16915471_16915622 | 0.46 |
Egfr |
epidermal growth factor receptor |
10361 |
0.18 |
| chr13_24889964_24890115 | 0.45 |
D130043K22Rik |
RIKEN cDNA D130043K22 gene |
3042 |
0.18 |
| chr1_67150788_67150939 | 0.45 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
27837 |
0.2 |
| chr1_162878367_162878530 | 0.45 |
Fmo2 |
flavin containing monooxygenase 2 |
2283 |
0.28 |
| chr15_3470264_3470480 | 0.45 |
Ghr |
growth hormone receptor |
1272 |
0.6 |
| chr6_94083257_94083610 | 0.44 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
169743 |
0.03 |
| chr5_131533929_131534082 | 0.44 |
Auts2 |
autism susceptibility candidate 2 |
392 |
0.86 |
| chr10_68090841_68090992 | 0.44 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
45710 |
0.14 |
| chr17_77735813_77735964 | 0.44 |
Gm4711 |
predicted gene 4711 |
48539 |
0.17 |
| chr16_24103845_24103996 | 0.43 |
Gm31583 |
predicted gene, 31583 |
13831 |
0.17 |
| chr16_77461400_77461565 | 0.43 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
4439 |
0.14 |
| chr3_138375535_138375764 | 0.43 |
Adh6-ps1 |
alcohol dehydrogenase 6 (class V), pseudogene 1 |
1459 |
0.3 |
| chr19_26778570_26778772 | 0.43 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
7075 |
0.21 |
| chr19_32509968_32510223 | 0.42 |
Minpp1 |
multiple inositol polyphosphate histidine phosphatase 1 |
24172 |
0.13 |
| chr3_24595827_24596029 | 0.42 |
Gm24704 |
predicted gene, 24704 |
33854 |
0.24 |
| chr2_138258025_138258203 | 0.42 |
Btbd3 |
BTB (POZ) domain containing 3 |
1444 |
0.6 |
| chr15_38115472_38115623 | 0.42 |
Gm49312 |
predicted gene, 49312 |
36445 |
0.12 |
| chr7_114788616_114788767 | 0.41 |
Insc |
INSC spindle orientation adaptor protein |
3362 |
0.24 |
| chr18_76254862_76255034 | 0.40 |
Smad2 |
SMAD family member 2 |
6173 |
0.2 |
| chr8_105811176_105811604 | 0.40 |
Ranbp10 |
RAN binding protein 10 |
15815 |
0.09 |
| chr18_69659921_69660072 | 0.40 |
Tcf4 |
transcription factor 4 |
3329 |
0.36 |
| chr17_24861821_24861975 | 0.40 |
Hagh |
hydroxyacyl glutathione hydrolase |
3697 |
0.1 |
| chr15_36592351_36592502 | 0.39 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
5000 |
0.15 |
| chr17_88304125_88304285 | 0.39 |
1700116H05Rik |
RIKEN cDNA 1700116H05 gene |
1173 |
0.51 |
| chr9_29643377_29643540 | 0.39 |
Gm15521 |
predicted gene 15521 |
50948 |
0.19 |
| chr2_152733626_152733777 | 0.39 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
2550 |
0.16 |
| chr14_17728323_17728474 | 0.38 |
Gm48320 |
predicted gene, 48320 |
42724 |
0.19 |
| chr9_55324808_55324982 | 0.38 |
Nrg4 |
neuregulin 4 |
292 |
0.88 |
| chr15_59430473_59430639 | 0.38 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
56269 |
0.12 |
| chr14_21181534_21181685 | 0.38 |
Adk |
adenosine kinase |
105457 |
0.06 |
| chr3_116204045_116204241 | 0.38 |
Gm31651 |
predicted gene, 31651 |
3735 |
0.2 |
| chr3_138455131_138455312 | 0.38 |
Adh5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
816 |
0.51 |
| chr17_84480594_84480759 | 0.37 |
Thada |
thyroid adenoma associated |
14471 |
0.18 |
| chr9_57525829_57526000 | 0.37 |
Cox5a |
cytochrome c oxidase subunit 5A |
4581 |
0.1 |
| chr9_70271019_70271199 | 0.37 |
Myo1e |
myosin IE |
63741 |
0.11 |
| chr15_96686781_96687182 | 0.37 |
Slc38a2 |
solute carrier family 38, member 2 |
11710 |
0.16 |
| chr4_131890254_131890430 | 0.37 |
Srsf4 |
serine and arginine-rich splicing factor 4 |
5134 |
0.11 |
| chr11_50312756_50312907 | 0.36 |
Canx |
calnexin |
11841 |
0.12 |
| chr11_19830960_19831117 | 0.36 |
Gm12029 |
predicted gene 12029 |
47386 |
0.15 |
| chr6_124709647_124709812 | 0.36 |
Emg1 |
EMG1 N1-specific pseudouridine methyltransferase |
1316 |
0.16 |
| chr3_129606663_129606955 | 0.35 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
54429 |
0.09 |
| chr6_51720618_51721036 | 0.35 |
Gm38811 |
predicted gene, 38811 |
9746 |
0.26 |
| chr1_162571667_162571910 | 0.35 |
Vamp4 |
vesicle-associated membrane protein 4 |
562 |
0.74 |
| chr17_46026823_46026974 | 0.35 |
Vegfa |
vascular endothelial growth factor A |
2017 |
0.29 |
| chr4_3318885_3319036 | 0.35 |
Gm11786 |
predicted gene 11786 |
8215 |
0.21 |
| chr11_75754732_75754970 | 0.35 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
3247 |
0.21 |
| chr6_100848852_100849036 | 0.34 |
Ppp4r2 |
protein phosphatase 4, regulatory subunit 2 |
3541 |
0.26 |
| chr19_56519861_56520029 | 0.34 |
Dclre1a |
DNA cross-link repair 1A |
17437 |
0.17 |
| chr7_46719930_46720314 | 0.34 |
Saa3 |
serum amyloid A 3 |
4422 |
0.1 |
| chr6_149147053_149147326 | 0.34 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
5543 |
0.14 |
| chr13_24144521_24144672 | 0.34 |
Carmil1 |
capping protein regulator and myosin 1 linker 1 |
10613 |
0.19 |
| chr10_87582564_87582726 | 0.33 |
Pah |
phenylalanine hydroxylase |
35972 |
0.17 |
| chr8_36319827_36320137 | 0.33 |
Gm5787 |
predicted gene 5787 |
53651 |
0.12 |
| chr1_13646788_13647045 | 0.33 |
Gm38319 |
predicted gene, 38319 |
4168 |
0.2 |
| chr5_103799401_103799738 | 0.33 |
Aff1 |
AF4/FMR2 family, member 1 |
15210 |
0.19 |
| chr2_59159714_59159865 | 0.33 |
Ccdc148 |
coiled-coil domain containing 148 |
745 |
0.51 |
| chr8_60947880_60948031 | 0.33 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
6793 |
0.17 |
| chr16_91829363_91829514 | 0.33 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
1901 |
0.33 |
| chr13_96795211_96795384 | 0.33 |
Ankrd31 |
ankyrin repeat domain 31 |
8701 |
0.18 |
| chr8_47708640_47708855 | 0.33 |
E030037K01Rik |
RIKEN cDNA E030037K01 gene |
4519 |
0.12 |
| chr13_43548363_43548529 | 0.33 |
Mcur1 |
mitochondrial calcium uniporter regulator 1 |
2326 |
0.24 |
| chr9_92251108_92251259 | 0.32 |
Plscr1 |
phospholipid scramblase 1 |
841 |
0.56 |
| chr11_83721035_83721187 | 0.32 |
Wfdc18 |
WAP four-disulfide core domain 18 |
11986 |
0.08 |
| chrX_42198771_42199059 | 0.32 |
Stag2 |
stromal antigen 2 |
46735 |
0.14 |
| chr6_91582630_91582782 | 0.32 |
Gm45215 |
predicted gene 45215 |
12052 |
0.14 |
| chr3_96562255_96562584 | 0.32 |
Txnip |
thioredoxin interacting protein |
2365 |
0.12 |
| chr4_6274068_6274223 | 0.32 |
Cyp7a1 |
cytochrome P450, family 7, subfamily a, polypeptide 1 |
1486 |
0.41 |
| chr11_113138785_113138971 | 0.32 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
34199 |
0.21 |
| chr13_81323962_81324134 | 0.32 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
18788 |
0.24 |
| chr5_87377364_87377538 | 0.32 |
Gm21049 |
predicted gene, 21049 |
2469 |
0.16 |
| chr6_17252773_17253114 | 0.31 |
Cav2 |
caveolin 2 |
28242 |
0.16 |
| chr16_93354892_93355097 | 0.31 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
892 |
0.57 |
| chr2_4929917_4930068 | 0.31 |
Phyh |
phytanoyl-CoA hydroxylase |
7491 |
0.13 |
| chr5_107369592_107369801 | 0.31 |
Brdt |
bromodomain, testis-specific |
9859 |
0.11 |
| chr12_59382932_59383083 | 0.31 |
Gm48268 |
predicted gene, 48268 |
27398 |
0.14 |
| chr19_56652542_56652727 | 0.31 |
Gm32441 |
predicted gene, 32441 |
11977 |
0.17 |
| chr5_123185860_123186059 | 0.31 |
Hpd |
4-hydroxyphenylpyruvic acid dioxygenase |
3232 |
0.12 |
| chr5_45278745_45278901 | 0.31 |
Gm43303 |
predicted gene 43303 |
26894 |
0.18 |
| chr17_78269187_78269367 | 0.31 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
10906 |
0.19 |
| chr6_119681279_119681450 | 0.31 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
59259 |
0.13 |
| chr18_76752705_76752870 | 0.31 |
Skor2 |
SKI family transcriptional corepressor 2 |
103618 |
0.07 |
| chr14_120355831_120355991 | 0.31 |
Mbnl2 |
muscleblind like splicing factor 2 |
23330 |
0.21 |
| chr1_156558196_156558347 | 0.31 |
Abl2 |
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
515 |
0.76 |
| chr13_91756942_91757255 | 0.31 |
Gm27656 |
predicted gene, 27656 |
4992 |
0.17 |
| chr9_111124362_111124513 | 0.30 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
4762 |
0.17 |
| chr1_107939897_107940125 | 0.30 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
3302 |
0.23 |
| chr2_58755277_58755512 | 0.30 |
Upp2 |
uridine phosphorylase 2 |
182 |
0.95 |
| chr15_97048203_97048354 | 0.30 |
Slc38a4 |
solute carrier family 38, member 4 |
1809 |
0.47 |
| chr4_105116678_105116829 | 0.30 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
6863 |
0.27 |
| chr15_66613033_66613221 | 0.30 |
Phf20l1 |
PHD finger protein 20-like 1 |
967 |
0.61 |
| chr5_89352355_89352713 | 0.30 |
Gc |
vitamin D binding protein |
83094 |
0.1 |
| chr3_14841128_14841305 | 0.30 |
Car3 |
carbonic anhydrase 3 |
22296 |
0.15 |
| chr6_120686819_120687199 | 0.30 |
Cecr2 |
CECR2, histone acetyl-lysine reader |
20427 |
0.18 |
| chr7_73549643_73549987 | 0.29 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
2950 |
0.15 |
| chr2_52613475_52613626 | 0.29 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
6204 |
0.24 |
| chr8_68060407_68060639 | 0.29 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
1704 |
0.46 |
| chrX_14897302_14897453 | 0.29 |
Gm9435 |
predicted gene 9435 |
58597 |
0.15 |
| chr11_77516276_77516445 | 0.29 |
Abhd15 |
abhydrolase domain containing 15 |
376 |
0.44 |
| chr13_113767020_113767171 | 0.29 |
Gm47463 |
predicted gene, 47463 |
2480 |
0.19 |
| chr19_8218872_8219037 | 0.29 |
Slc22a29 |
solute carrier family 22. member 29 |
54 |
0.97 |
| chr15_85766704_85766873 | 0.29 |
Ppara |
peroxisome proliferator activated receptor alpha |
5426 |
0.16 |
| chr10_19878984_19879150 | 0.29 |
Gm48714 |
predicted gene, 48714 |
4620 |
0.19 |
| chr2_27768705_27768856 | 0.28 |
Rxra |
retinoid X receptor alpha |
28579 |
0.21 |
| chr11_107473047_107473198 | 0.28 |
Pitpnc1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
2423 |
0.19 |
| chr9_110084221_110084372 | 0.27 |
Dhx30 |
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
695 |
0.58 |
| chr9_83576757_83576932 | 0.27 |
Sh3bgrl2 |
SH3 domain binding glutamic acid-rich protein like 2 |
13 |
0.98 |
| chr16_25967441_25967597 | 0.27 |
Gm4524 |
predicted gene 4524 |
11044 |
0.22 |
| chr12_35265470_35265829 | 0.27 |
Gm44396 |
predicted gene, 44396 |
43242 |
0.16 |
| chr7_73608284_73608633 | 0.27 |
Gm44734 |
predicted gene 44734 |
490 |
0.68 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.3 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.5 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.1 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.3 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.0 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.5 | GO:0052770 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0052767 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |