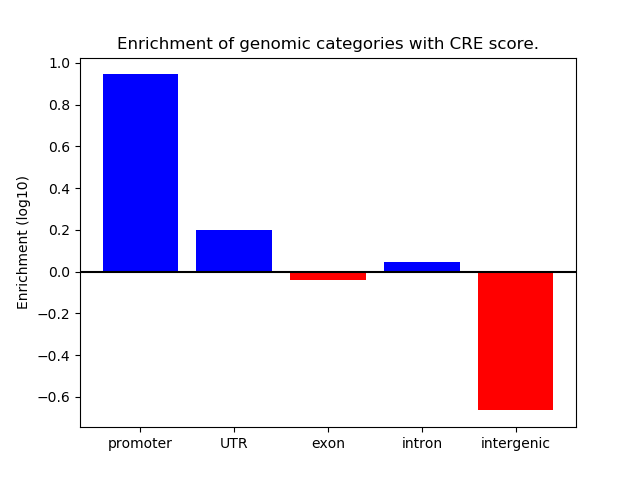
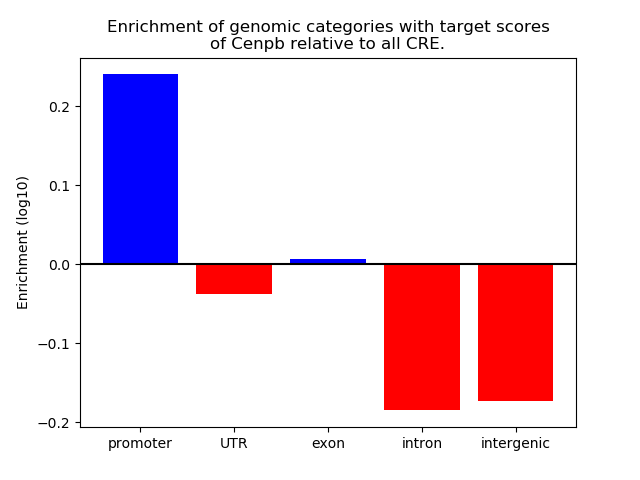
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
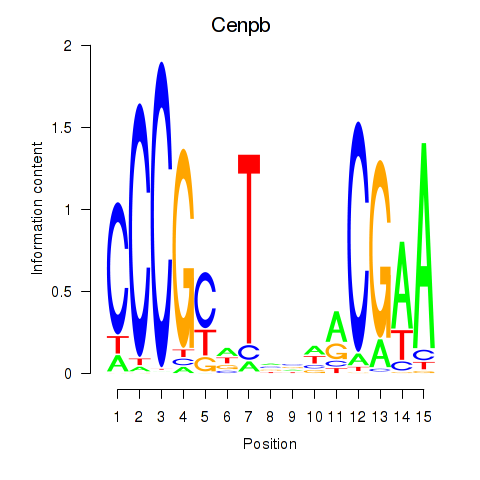
Results for Cenpb
Z-value: 1.41
Transcription factors associated with Cenpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cenpb
|
ENSMUSG00000068267.4 | centromere protein B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_131179083_131179346 | Cenpb | 853 | 0.400629 | 0.75 | 8.6e-02 | Click! |
| chr2_131179412_131179897 | Cenpb | 413 | 0.699909 | 0.70 | 1.2e-01 | Click! |
| chr2_131178818_131179001 | Cenpb | 1158 | 0.290260 | -0.18 | 7.4e-01 | Click! |
| chr2_131180305_131180466 | Cenpb | 318 | 0.782939 | 0.12 | 8.2e-01 | Click! |
| chr2_131180016_131180294 | Cenpb | 88 | 0.935173 | 0.08 | 8.7e-01 | Click! |
Activity of the Cenpb motif across conditions
Conditions sorted by the z-value of the Cenpb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_66816824_66817235 | 0.57 |
Kansl1l |
KAT8 regulatory NSL complex subunit 1-like |
517 |
0.58 |
| chrX_113094960_113095111 | 0.50 |
Mir361 |
microRNA 361 |
20142 |
0.18 |
| chr13_93632435_93632697 | 0.50 |
Bhmt |
betaine-homocysteine methyltransferase |
5000 |
0.17 |
| chr10_51721458_51721757 | 0.46 |
Rfx6 |
regulatory factor X, 6 |
27836 |
0.12 |
| chr8_124273183_124273568 | 0.45 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
21728 |
0.15 |
| chr10_81600790_81601143 | 0.45 |
Tle6 |
transducin-like enhancer of split 6 |
66 |
0.93 |
| chr1_72176192_72176343 | 0.44 |
Mreg |
melanoregulin |
36040 |
0.11 |
| chr2_181319647_181320163 | 0.44 |
Rtel1 |
regulator of telomere elongation helicase 1 |
58 |
0.95 |
| chr9_50844938_50845089 | 0.42 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
288 |
0.88 |
| chrX_36329032_36329187 | 0.39 |
Lonrf3 |
LON peptidase N-terminal domain and ring finger 3 |
107 |
0.96 |
| chr16_31948717_31948903 | 0.39 |
Ncbp2 |
nuclear cap binding protein subunit 2 |
164 |
0.65 |
| chr9_107986066_107986266 | 0.38 |
Inka1 |
inka box actin regulator 1 |
287 |
0.67 |
| chr2_118487263_118487442 | 0.38 |
Gm13983 |
predicted gene 13983 |
3175 |
0.2 |
| chr17_28517484_28517972 | 0.38 |
Fkbp5 |
FK506 binding protein 5 |
201 |
0.84 |
| chr8_35826198_35826406 | 0.38 |
Cldn23 |
claudin 23 |
257 |
0.91 |
| chr13_93622998_93623302 | 0.37 |
Gm15622 |
predicted gene 15622 |
2232 |
0.26 |
| chr14_64679206_64679357 | 0.36 |
Kif13b |
kinesin family member 13B |
26679 |
0.17 |
| chr17_71662488_71662673 | 0.36 |
Wdr43 |
WD repeat domain 43 |
5484 |
0.11 |
| chr11_75167957_75168108 | 0.35 |
Hic1 |
hypermethylated in cancer 1 |
114 |
0.92 |
| chr8_85606671_85606835 | 0.33 |
Gm45425 |
predicted gene 45425 |
1291 |
0.4 |
| chr10_22361736_22361909 | 0.33 |
Raet1d |
retinoic acid early transcript delta |
72 |
0.96 |
| chr2_34882484_34882704 | 0.33 |
Psmd5 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
7626 |
0.11 |
| chr1_138617784_138617983 | 0.32 |
Nek7 |
NIMA (never in mitosis gene a)-related expressed kinase 7 |
1791 |
0.35 |
| chr14_55897498_55897649 | 0.32 |
Sdr39u1 |
short chain dehydrogenase/reductase family 39U, member 1 |
2282 |
0.15 |
| chr19_61226929_61227456 | 0.31 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr9_43808674_43808844 | 0.31 |
Nectin1 |
nectin cell adhesion molecule 1 |
12188 |
0.18 |
| chr2_167843799_167844308 | 0.31 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
10407 |
0.16 |
| chr15_99126293_99126465 | 0.30 |
Spats2 |
spermatogenesis associated, serine-rich 2 |
177 |
0.9 |
| chr1_21261382_21261702 | 0.30 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8021 |
0.11 |
| chr1_21249667_21249920 | 0.30 |
Gsta3 |
glutathione S-transferase, alpha 3 |
3728 |
0.14 |
| chr7_19629444_19629613 | 0.30 |
Relb |
avian reticuloendotheliosis viral (v-rel) oncogene related B |
90 |
0.93 |
| chr5_120490437_120490593 | 0.30 |
Gm15690 |
predicted gene 15690 |
9612 |
0.1 |
| chr2_172720968_172721136 | 0.29 |
Gm22773 |
predicted gene, 22773 |
143289 |
0.04 |
| chr17_56105602_56105761 | 0.29 |
Plin4 |
perilipin 4 |
4121 |
0.1 |
| chr10_22173370_22173547 | 0.29 |
Raet1e |
retinoic acid early transcript 1E |
63 |
0.95 |
| chr11_87968578_87968738 | 0.29 |
Dynll2 |
dynein light chain LC8-type 2 |
15354 |
0.11 |
| chr7_25687287_25687542 | 0.29 |
Tgfb1 |
transforming growth factor, beta 1 |
412 |
0.69 |
| chr16_30520108_30520279 | 0.29 |
Tmem44 |
transmembrane protein 44 |
20716 |
0.18 |
| chr7_64802263_64802423 | 0.29 |
Fam189a1 |
family with sequence similarity 189, member A1 |
28691 |
0.19 |
| chr11_52099511_52099749 | 0.28 |
Gm24157 |
predicted gene, 24157 |
477 |
0.61 |
| chr3_108778882_108779033 | 0.28 |
Aknad1 |
AKNA domain containing 1 |
18743 |
0.13 |
| chr7_28378113_28378305 | 0.28 |
Gm44710 |
predicted gene 44710 |
771 |
0.32 |
| chr6_72565635_72565786 | 0.28 |
Capg |
capping protein (actin filament), gelsolin-like |
9731 |
0.1 |
| chr2_28840041_28840192 | 0.28 |
Gtf3c4 |
general transcription factor IIIC, polypeptide 4 |
170 |
0.65 |
| chr9_48753826_48754020 | 0.28 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
82022 |
0.09 |
| chr8_61650614_61650765 | 0.28 |
Palld |
palladin, cytoskeletal associated protein |
59547 |
0.14 |
| chr3_86070700_86070889 | 0.27 |
Sh3d19 |
SH3 domain protein D19 |
121 |
0.95 |
| chr6_124683057_124683263 | 0.27 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
18259 |
0.06 |
| chr7_143769241_143769509 | 0.27 |
Mrgprg |
MAS-related GPR, member G |
2382 |
0.17 |
| chr9_55541381_55541949 | 0.27 |
Isl2 |
insulin related protein 2 (islet 2) |
458 |
0.61 |
| chr11_97575545_97575704 | 0.27 |
Srcin1 |
SRC kinase signaling inhibitor 1 |
398 |
0.78 |
| chr15_57695893_57696526 | 0.27 |
Zhx2 |
zinc fingers and homeoboxes 2 |
480 |
0.82 |
| chr15_82206081_82206267 | 0.27 |
Shisa8 |
shisa family member 8 |
3766 |
0.1 |
| chr10_122449019_122449793 | 0.26 |
Avpr1a |
arginine vasopressin receptor 1A |
907 |
0.59 |
| chr6_128362234_128363003 | 0.26 |
Rhno1 |
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
39 |
0.8 |
| chr19_36909759_36909919 | 0.26 |
Fgfbp3 |
fibroblast growth factor binding protein 3 |
9760 |
0.18 |
| chr1_58796952_58797103 | 0.26 |
Gm37760 |
predicted gene, 37760 |
756 |
0.51 |
| chr10_42229040_42229314 | 0.26 |
Foxo3 |
forkhead box O3 |
29189 |
0.21 |
| chr9_65827242_65827393 | 0.26 |
Zfp609 |
zinc finger protein 609 |
247 |
0.89 |
| chr11_121057893_121058050 | 0.26 |
Sectm1b |
secreted and transmembrane 1B |
1343 |
0.24 |
| chr3_97226539_97226936 | 0.26 |
Bcl9 |
B cell CLL/lymphoma 9 |
627 |
0.7 |
| chr6_115037407_115037571 | 0.26 |
Tamm41 |
TAM41 mitochondrial translocator assembly and maintenance homolog |
336 |
0.89 |
| chr3_84228289_84228569 | 0.26 |
Trim2 |
tripartite motif-containing 2 |
7540 |
0.24 |
| chr1_133710698_133710864 | 0.25 |
Atp2b4 |
ATPase, Ca++ transporting, plasma membrane 4 |
7148 |
0.14 |
| chr15_79455529_79455727 | 0.25 |
Csnk1e |
casein kinase 1, epsilon |
62 |
0.95 |
| chr13_55693653_55693804 | 0.25 |
B230219D22Rik |
RIKEN cDNA B230219D22 gene |
604 |
0.63 |
| chr14_26669361_26669540 | 0.25 |
Mir7672 |
microRNA 7672 |
158 |
0.69 |
| chr12_17691350_17691514 | 0.25 |
Hpcal1 |
hippocalcin-like 1 |
576 |
0.8 |
| chr14_63248471_63248829 | 0.25 |
Gata4 |
GATA binding protein 4 |
3379 |
0.22 |
| chr14_31195860_31196041 | 0.25 |
Nisch |
nischarin |
3331 |
0.14 |
| chr14_55659957_55660132 | 0.25 |
Mdp1 |
magnesium-dependent phosphatase 1 |
386 |
0.56 |
| chr15_102186991_102187189 | 0.24 |
Csad |
cysteine sulfinic acid decarboxylase |
509 |
0.65 |
| chr5_114553572_114553723 | 0.24 |
Gm13790 |
predicted gene 13790 |
2358 |
0.25 |
| chr6_124688048_124688642 | 0.24 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
13074 |
0.06 |
| chr2_119787882_119788047 | 0.24 |
Rpap1 |
RNA polymerase II associated protein 1 |
427 |
0.73 |
| chr8_119925715_119925866 | 0.24 |
Usp10 |
ubiquitin specific peptidase 10 |
11892 |
0.16 |
| chr19_46056737_46057343 | 0.24 |
Pprc1 |
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
176 |
0.92 |
| chr2_170511584_170512109 | 0.24 |
Gm16796 |
predicted gene, 16796 |
81 |
0.76 |
| chr11_84523271_84523458 | 0.24 |
Lhx1 |
LIM homeobox protein 1 |
653 |
0.67 |
| chr14_60980883_60981065 | 0.24 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
5089 |
0.27 |
| chr17_45700190_45700435 | 0.24 |
Mrpl14 |
mitochondrial ribosomal protein L14 |
4896 |
0.13 |
| chr17_31901765_31901975 | 0.24 |
Gm50000 |
predicted gene, 50000 |
2692 |
0.2 |
| chr13_112466009_112466408 | 0.24 |
Il6st |
interleukin 6 signal transducer |
1385 |
0.38 |
| chr17_56462880_56463031 | 0.24 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
739 |
0.6 |
| chr11_90686948_90687471 | 0.24 |
Tom1l1 |
target of myb1-like 1 (chicken) |
370 |
0.89 |
| chr18_68260862_68261044 | 0.24 |
Mir7219 |
microRNA 7219 |
20 |
0.98 |
| chr19_54206776_54207130 | 0.24 |
Gm50187 |
predicted gene, 50187 |
8562 |
0.28 |
| chr13_93619970_93620121 | 0.23 |
Gm15622 |
predicted gene 15622 |
5337 |
0.18 |
| chr2_126424829_126424981 | 0.23 |
Atp8b4 |
ATPase, class I, type 8B, member 4 |
66648 |
0.12 |
| chr10_78287868_78288030 | 0.23 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
76 |
0.94 |
| chr12_77239645_77239931 | 0.23 |
Fut8 |
fucosyltransferase 8 |
723 |
0.56 |
| chr4_44976602_44976791 | 0.23 |
Gm12679 |
predicted gene 12679 |
1623 |
0.24 |
| chr11_11942496_11942647 | 0.23 |
Grb10 |
growth factor receptor bound protein 10 |
3759 |
0.24 |
| chr13_51072573_51072833 | 0.23 |
Spin1 |
spindlin 1 |
28177 |
0.2 |
| chr4_132884617_132884863 | 0.23 |
Gm13033 |
predicted gene 13033 |
23 |
0.75 |
| chr6_39257305_39257499 | 0.23 |
Gm26008 |
predicted gene, 26008 |
8540 |
0.16 |
| chr12_104854088_104854239 | 0.23 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
3208 |
0.23 |
| chr19_16133750_16134171 | 0.23 |
Gnaq |
guanine nucleotide binding protein, alpha q polypeptide |
664 |
0.74 |
| chr2_180589339_180589504 | 0.23 |
Ogfr |
opioid growth factor receptor |
41 |
0.96 |
| chr18_39693461_39693788 | 0.23 |
Gm50398 |
predicted gene, 50398 |
32747 |
0.16 |
| chr6_131387426_131387577 | 0.23 |
Ybx3 |
Y box protein 3 |
937 |
0.48 |
| chr1_21344197_21344382 | 0.23 |
Khdc1a |
KH domain containing 1A |
5352 |
0.1 |
| chr9_64087364_64087541 | 0.23 |
Scarletltr |
Scarletltr, erythroid developmental long intergenic non-protein coding transcript |
6691 |
0.13 |
| chr8_122678475_122678654 | 0.23 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
130 |
0.93 |
| chr5_63886612_63886794 | 0.23 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
9227 |
0.19 |
| chr14_14346051_14346426 | 0.22 |
Il3ra |
interleukin 3 receptor, alpha chain |
37 |
0.94 |
| chr6_72601356_72601528 | 0.22 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
2814 |
0.11 |
| chr12_83987263_83987451 | 0.22 |
Acot2 |
acyl-CoA thioesterase 2 |
504 |
0.62 |
| chr19_55892938_55893099 | 0.22 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
334 |
0.92 |
| chr9_57552287_57552535 | 0.22 |
Mpi |
mannose phosphate isomerase |
145 |
0.91 |
| chr14_114856480_114856899 | 0.22 |
Gm49010 |
predicted gene, 49010 |
19821 |
0.16 |
| chr1_40156718_40157115 | 0.22 |
Il1r1 |
interleukin 1 receptor, type I |
68164 |
0.08 |
| chr1_135146963_135147134 | 0.22 |
Arl8a |
ADP-ribosylation factor-like 8A |
224 |
0.86 |
| chr5_122268614_122268852 | 0.22 |
Tctn1 |
tectonic family member 1 |
4273 |
0.14 |
| chr6_113891230_113891419 | 0.22 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
46 |
0.98 |
| chr11_69902083_69902235 | 0.22 |
Neurl4 |
neuralized E3 ubiquitin protein ligase 4 |
3 |
0.9 |
| chr4_150754056_150754234 | 0.22 |
Gm13049 |
predicted gene 13049 |
71588 |
0.08 |
| chr5_140103811_140104157 | 0.21 |
Gm16121 |
predicted gene 16121 |
2785 |
0.22 |
| chr2_71212098_71212533 | 0.21 |
Dync1i2 |
dynein cytoplasmic 1 intermediate chain 2 |
311 |
0.92 |
| chr9_44478965_44479362 | 0.21 |
C030014I23Rik |
RIKEN cDNA C030014I23 gene |
385 |
0.49 |
| chr3_14533935_14534109 | 0.21 |
Lrrcc1 |
leucine rich repeat and coiled-coil domain containing 1 |
88 |
0.96 |
| chr12_79436402_79436553 | 0.21 |
Rad51b |
RAD51 paralog B |
109124 |
0.06 |
| chr1_169919728_169919879 | 0.21 |
Ccdc190 |
coiled-coil domain containing 190 |
8845 |
0.17 |
| chr19_17394223_17394397 | 0.21 |
Rfk |
riboflavin kinase |
258 |
0.94 |
| chr7_109752423_109752599 | 0.21 |
4930431P19Rik |
RIKEN cDNA 4930431P19 gene |
191 |
0.53 |
| chr15_36471395_36471566 | 0.21 |
Ankrd46 |
ankyrin repeat domain 46 |
25235 |
0.12 |
| chr2_127007135_127007298 | 0.21 |
Ap4e1 |
adaptor-related protein complex AP-4, epsilon 1 |
1501 |
0.38 |
| chr4_101420987_101421138 | 0.21 |
Ak4 |
adenylate kinase 4 |
565 |
0.7 |
| chr16_92610894_92611062 | 0.21 |
Gm26626 |
predicted gene, 26626 |
1836 |
0.39 |
| chr7_27731043_27731227 | 0.21 |
Zfp60 |
zinc finger protein 60 |
254 |
0.88 |
| chr8_70775984_70776205 | 0.21 |
Pik3r2 |
phosphoinositide-3-kinase regulatory subunit 2 |
585 |
0.36 |
| chr15_102181335_102181499 | 0.20 |
Csad |
cysteine sulfinic acid decarboxylase |
1556 |
0.24 |
| chr2_92055713_92056044 | 0.20 |
2900072N19Rik |
RIKEN cDNA 2900072N19 gene |
3491 |
0.19 |
| chr13_93634275_93634633 | 0.20 |
Bhmt |
betaine-homocysteine methyltransferase |
3112 |
0.2 |
| chr4_15319290_15319441 | 0.20 |
Tmem64 |
transmembrane protein 64 |
53534 |
0.14 |
| chr1_172563069_172563473 | 0.20 |
Vsig8 |
V-set and immunoglobulin domain containing 8 |
3365 |
0.14 |
| chr5_122799376_122799527 | 0.20 |
Anapc5 |
anaphase-promoting complex subunit 5 |
279 |
0.87 |
| chr1_194977127_194977294 | 0.20 |
Gm16897 |
predicted gene, 16897 |
250 |
0.78 |
| chr17_29031946_29032467 | 0.20 |
Gm16196 |
predicted gene 16196 |
362 |
0.47 |
| chr4_44991523_44991952 | 0.20 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
10237 |
0.11 |
| chr15_58415328_58415520 | 0.20 |
Fam91a1 |
family with sequence similarity 91, member A1 |
44 |
0.97 |
| chr1_93224076_93224260 | 0.20 |
Sned1 |
sushi, nidogen and EGF-like domains 1 |
11673 |
0.13 |
| chr12_100935147_100935320 | 0.20 |
Ccdc88c |
coiled-coil domain containing 88C |
6168 |
0.13 |
| chr4_136135254_136135405 | 0.20 |
Gm26001 |
predicted gene, 26001 |
2167 |
0.23 |
| chr10_108435314_108435492 | 0.20 |
Gm36283 |
predicted gene, 36283 |
1262 |
0.46 |
| chr19_46138051_46138212 | 0.20 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
842 |
0.48 |
| chr4_12906927_12907078 | 0.20 |
Triqk |
triple QxxK/R motif containing |
144 |
0.98 |
| chr10_51722569_51722927 | 0.20 |
Rfx6 |
regulatory factor X, 6 |
28977 |
0.12 |
| chr8_122678735_122678891 | 0.20 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
119 |
0.93 |
| chr7_81542702_81542853 | 0.20 |
Gm37829 |
predicted gene, 37829 |
10256 |
0.1 |
| chr4_133039253_133039404 | 0.20 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
165 |
0.95 |
| chr19_4962457_4962761 | 0.20 |
Mrpl11 |
mitochondrial ribosomal protein L11 |
235 |
0.83 |
| chr3_122729656_122729846 | 0.20 |
Pde5a |
phosphodiesterase 5A, cGMP-specific |
181 |
0.95 |
| chr8_22860131_22860751 | 0.19 |
Kat6a |
K(lysine) acetyltransferase 6A |
892 |
0.52 |
| chr10_127066593_127066777 | 0.19 |
Cdk4 |
cyclin-dependent kinase 4 |
609 |
0.45 |
| chr1_36556917_36557103 | 0.19 |
Sema4c |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
516 |
0.45 |
| chr13_108270139_108270307 | 0.19 |
Gm30747 |
predicted gene, 30747 |
36668 |
0.11 |
| chr11_40692132_40692292 | 0.19 |
Mat2b |
methionine adenosyltransferase II, beta |
409 |
0.83 |
| chr13_55471689_55471869 | 0.19 |
Mir6944 |
microRNA 6944 |
5997 |
0.08 |
| chr11_117076926_117077274 | 0.19 |
Gm26083 |
predicted gene, 26083 |
143 |
0.5 |
| chr11_70653511_70654114 | 0.19 |
Pfn1 |
profilin 1 |
786 |
0.31 |
| chr8_84907848_84907999 | 0.19 |
Dnase2a |
deoxyribonuclease II alpha |
637 |
0.43 |
| chr9_48984872_48985421 | 0.19 |
Usp28 |
ubiquitin specific peptidase 28 |
229 |
0.92 |
| chr10_18318743_18319176 | 0.19 |
Nhsl1 |
NHS-like 1 |
26 |
0.98 |
| chr9_69392079_69392263 | 0.19 |
Ice2 |
interactor of little elongation complex ELL subunit 2 |
5735 |
0.17 |
| chr18_64333176_64333751 | 0.19 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
243 |
0.91 |
| chr7_43562088_43562260 | 0.19 |
Zfp658 |
zinc finger protein 658 |
82 |
0.9 |
| chr9_50659707_50659907 | 0.19 |
Dlat |
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
27 |
0.96 |
| chr11_100935727_100935878 | 0.19 |
Stat3 |
signal transducer and activator of transcription 3 |
3578 |
0.18 |
| chr10_123841631_123841817 | 0.19 |
Gm18510 |
predicted gene, 18510 |
50174 |
0.19 |
| chr1_120265510_120265742 | 0.19 |
Steap3 |
STEAP family member 3 |
342 |
0.91 |
| chr10_77083914_77084160 | 0.19 |
Col18a1 |
collagen, type XVIII, alpha 1 |
5391 |
0.18 |
| chr4_143192114_143192277 | 0.19 |
Prdm2 |
PR domain containing 2, with ZNF domain |
2540 |
0.21 |
| chr14_25637635_25637888 | 0.19 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
19069 |
0.16 |
| chr19_42046010_42046180 | 0.19 |
Hoga1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
244 |
0.86 |
| chr6_148945212_148945706 | 0.19 |
3010003L21Rik |
RIKEN cDNA 3010003L21 gene |
592 |
0.4 |
| chr6_117907668_117908056 | 0.18 |
4933440N22Rik |
RIKEN cDNA 4933440N22 gene |
102 |
0.91 |
| chr17_31939698_31939868 | 0.18 |
Gm17572 |
predicted gene, 17572 |
10751 |
0.14 |
| chr5_38503360_38503605 | 0.18 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
339 |
0.86 |
| chr18_64641731_64641886 | 0.18 |
Atp8b1 |
ATPase, class I, type 8B, member 1 |
18999 |
0.13 |
| chr14_61680870_61681697 | 0.18 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr2_36074102_36074298 | 0.18 |
Gm43764 |
predicted gene 43764 |
4911 |
0.15 |
| chr12_73861884_73862127 | 0.18 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr7_63896677_63896828 | 0.18 |
Gm27252 |
predicted gene 27252 |
219 |
0.91 |
| chr6_88445522_88446347 | 0.18 |
Gm44264 |
predicted gene, 44264 |
247 |
0.73 |
| chr15_6709626_6709854 | 0.18 |
Rictor |
RPTOR independent companion of MTOR, complex 2 |
1357 |
0.47 |
| chr16_58523081_58523364 | 0.18 |
St3gal6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
122 |
0.96 |
| chr4_95881579_95881746 | 0.18 |
Fggy |
FGGY carbohydrate kinase domain containing |
32037 |
0.19 |
| chr10_128411776_128411940 | 0.18 |
Rnf41 |
ring finger protein 41 |
112 |
0.74 |
| chr10_20150520_20150679 | 0.18 |
Map7 |
microtubule-associated protein 7 |
1601 |
0.37 |
| chr17_35089633_35089905 | 0.18 |
Abhd16a |
abhydrolase domain containing 16A |
101 |
0.86 |
| chr9_111118266_111118417 | 0.18 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
104 |
0.96 |
| chr1_170880172_170880851 | 0.18 |
Dusp12 |
dual specificity phosphatase 12 |
4493 |
0.11 |
| chr11_94388312_94388483 | 0.18 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
4523 |
0.18 |
| chr18_20974383_20974578 | 0.18 |
Rnf125 |
ring finger protein 125 |
13008 |
0.2 |
| chr6_108665497_108666008 | 0.18 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
2706 |
0.23 |
| chr12_46819464_46819646 | 0.18 |
Nova1 |
NOVA alternative splicing regulator 1 |
626 |
0.75 |
| chr1_136933160_136933350 | 0.18 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
7328 |
0.24 |
| chr14_25726844_25727050 | 0.18 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
7155 |
0.15 |
| chr8_11758002_11758195 | 0.18 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
219 |
0.9 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.2 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 0.3 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.2 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.2 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.3 | GO:0003284 | septum primum development(GO:0003284) |
| 0.1 | 0.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.3 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.1 | 0.3 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.3 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.0 | GO:2000407 | regulation of T cell extravasation(GO:2000407) positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:1902430 | negative regulation of beta-amyloid formation(GO:1902430) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.0 | 0.2 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:1902992 | negative regulation of amyloid precursor protein catabolic process(GO:1902992) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0098597 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.0 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0051095 | regulation of helicase activity(GO:0051095) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.0 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.0 | GO:0035246 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine N-methylation(GO:0035246) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.0 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.0 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.3 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |