Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
Results for Clock
Z-value: 0.95
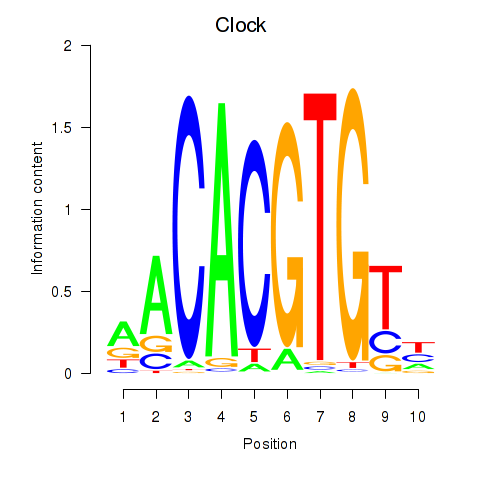
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSMUSG00000029238.8 | circadian locomotor output cycles kaput |
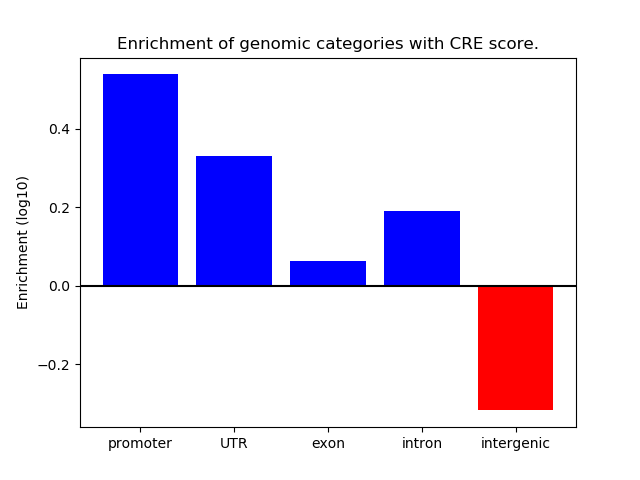
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_76304107_76304258 | Clock | 366 | 0.632461 | -0.87 | 2.3e-02 | Click! |
| chr5_76303540_76303814 | Clock | 871 | 0.362384 | -0.79 | 6.4e-02 | Click! |
| chr5_76303922_76304075 | Clock | 550 | 0.511694 | -0.75 | 8.5e-02 | Click! |
| chr5_76304752_76304909 | Clock | 38 | 0.497417 | -0.61 | 2.0e-01 | Click! |
| chr5_76256521_76256672 | Clock | 17416 | 0.123511 | 0.44 | 3.8e-01 | Click! |
Activity of the Clock motif across conditions
Conditions sorted by the z-value of the Clock motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
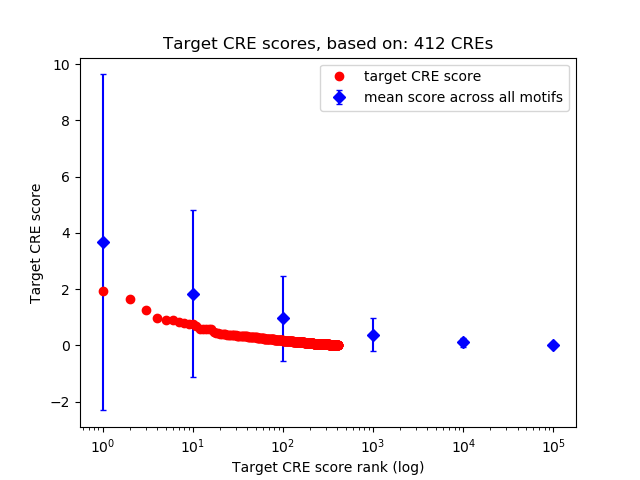
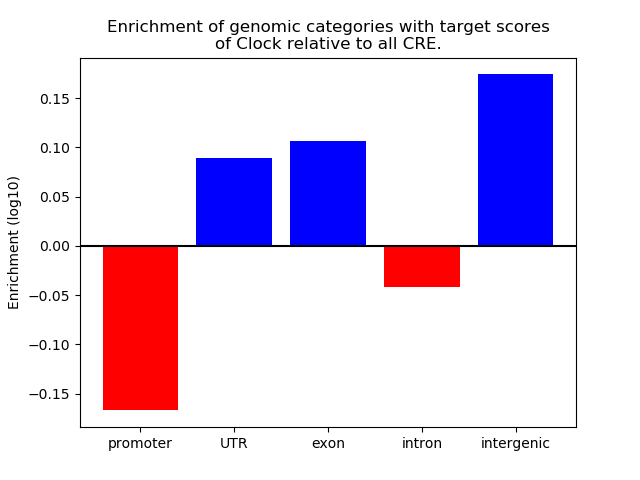
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_124005727_124005878 | 1.93 |
Gm12902 |
predicted gene 12902 |
79568 |
0.07 |
| chr16_48765285_48765461 | 1.64 |
Trat1 |
T cell receptor associated transmembrane adaptor 1 |
6583 |
0.2 |
| chr9_74331308_74331508 | 1.27 |
Gm24141 |
predicted gene, 24141 |
31202 |
0.18 |
| chr11_118265609_118265912 | 0.98 |
Usp36 |
ubiquitin specific peptidase 36 |
288 |
0.89 |
| chr19_33489637_33489788 | 0.89 |
Lipo5 |
lipase, member O5 |
16521 |
0.14 |
| chr19_33480626_33480777 | 0.88 |
Lipo5 |
lipase, member O5 |
7510 |
0.16 |
| chr1_186358132_186358570 | 0.82 |
Gm37491 |
predicted gene, 37491 |
11036 |
0.28 |
| chr17_71616884_71617050 | 0.78 |
Wdr43 |
WD repeat domain 43 |
752 |
0.37 |
| chr14_20479878_20480231 | 0.75 |
Anxa7 |
annexin A7 |
67 |
0.96 |
| chr4_101176228_101176401 | 0.75 |
Jak1 |
Janus kinase 1 |
9111 |
0.15 |
| chr5_8157542_8157699 | 0.68 |
Gm21759 |
predicted gene, 21759 |
22016 |
0.16 |
| chr10_95152509_95152710 | 0.58 |
Gm29684 |
predicted gene, 29684 |
6784 |
0.16 |
| chr6_148833285_148833458 | 0.58 |
Ipo8 |
importin 8 |
1904 |
0.22 |
| chr5_54060085_54060253 | 0.58 |
Stim2 |
stromal interaction molecule 2 |
14568 |
0.23 |
| chr18_60922022_60922173 | 0.57 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
3521 |
0.17 |
| chr3_57435352_57435577 | 0.56 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
10150 |
0.22 |
| chr7_68311256_68311435 | 0.47 |
4930405G09Rik |
RIKEN cDNA 4930405G09 gene |
5687 |
0.14 |
| chr18_38406873_38407073 | 0.44 |
Ndfip1 |
Nedd4 family interacting protein 1 |
3423 |
0.18 |
| chr3_96092507_96092658 | 0.43 |
Gm43554 |
predicted gene 43554 |
5711 |
0.11 |
| chr19_37433970_37434137 | 0.41 |
Gm38345 |
predicted gene, 38345 |
603 |
0.45 |
| chr10_89484240_89484407 | 0.41 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
22326 |
0.19 |
| chrX_52044793_52044944 | 0.39 |
1700080O16Rik |
RIKEN cDNA 1700080O16 gene |
72096 |
0.11 |
| chr3_65663297_65663473 | 0.39 |
Lekr1 |
leucine, glutamate and lysine rich 1 |
2843 |
0.19 |
| chr2_156065132_156065865 | 0.37 |
Spag4 |
sperm associated antigen 4 |
55 |
0.94 |
| chr11_61974443_61974594 | 0.37 |
Gm12274 |
predicted gene 12274 |
168 |
0.92 |
| chr14_21832226_21832384 | 0.37 |
Vdac2 |
voltage-dependent anion channel 2 |
142 |
0.94 |
| chr11_61092676_61092849 | 0.37 |
Kcnj12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
26958 |
0.13 |
| chr9_107674967_107675413 | 0.36 |
Gnat1 |
guanine nucleotide binding protein, alpha transducing 1 |
1251 |
0.24 |
| chr4_63268906_63269081 | 0.35 |
Mir455 |
microRNA 455 |
12142 |
0.16 |
| chr6_50526613_50527019 | 0.35 |
Gm26289 |
predicted gene, 26289 |
13218 |
0.13 |
| chr11_44518397_44518764 | 0.35 |
Rnf145 |
ring finger protein 145 |
384 |
0.85 |
| chr11_120811562_120811973 | 0.35 |
Fasn |
fatty acid synthase |
2066 |
0.16 |
| chr10_96778362_96778523 | 0.34 |
Gm6859 |
predicted gene 6859 |
8614 |
0.17 |
| chr6_28895214_28895400 | 0.34 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
15264 |
0.21 |
| chr18_51128419_51128586 | 0.34 |
Prr16 |
proline rich 16 |
10764 |
0.3 |
| chr7_144940237_144940434 | 0.33 |
Ccnd1 |
cyclin D1 |
410 |
0.78 |
| chr1_131716457_131716608 | 0.32 |
Rab7b |
RAB7B, member RAS oncogene family |
19894 |
0.14 |
| chr19_26764962_26765268 | 0.32 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
5376 |
0.24 |
| chr8_46216985_46217176 | 0.31 |
Slc25a4 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
5796 |
0.11 |
| chr2_134486534_134486685 | 0.31 |
Hao1 |
hydroxyacid oxidase 1, liver |
67698 |
0.14 |
| chr5_87373500_87373711 | 0.31 |
Gm35570 |
predicted gene, 35570 |
3122 |
0.14 |
| chr14_69729450_69729663 | 0.31 |
Chmp7 |
charged multivesicular body protein 7 |
1319 |
0.33 |
| chr7_79363538_79363689 | 0.31 |
Rlbp1 |
retinaldehyde binding protein 1 |
14090 |
0.13 |
| chr11_5912275_5912428 | 0.30 |
Gck |
glucokinase |
2773 |
0.16 |
| chr6_117577945_117578096 | 0.29 |
Gm9946 |
predicted gene 9946 |
10348 |
0.2 |
| chr4_150218378_150218529 | 0.29 |
Gm13094 |
predicted gene 13094 |
9582 |
0.13 |
| chr13_63665698_63665976 | 0.29 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr19_40417273_40417445 | 0.29 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
15092 |
0.22 |
| chr16_43552061_43552212 | 0.29 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
41828 |
0.16 |
| chr11_82845122_82845302 | 0.28 |
Rffl |
ring finger and FYVE like domain containing protein |
880 |
0.45 |
| chr8_109681119_109681276 | 0.28 |
Ist1 |
increased sodium tolerance 1 homolog (yeast) |
3603 |
0.19 |
| chr4_117981260_117981475 | 0.27 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
6331 |
0.16 |
| chr10_19443595_19443775 | 0.27 |
Gm33104 |
predicted gene, 33104 |
43030 |
0.15 |
| chr3_27283500_27283672 | 0.26 |
Gm37191 |
predicted gene, 37191 |
5526 |
0.2 |
| chr10_62647927_62648078 | 0.26 |
Ddx50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
221 |
0.89 |
| chr15_93628063_93628509 | 0.26 |
Prickle1 |
prickle planar cell polarity protein 1 |
32395 |
0.18 |
| chr1_88316757_88316908 | 0.25 |
Trpm8 |
transient receptor potential cation channel, subfamily M, member 8 |
2081 |
0.23 |
| chr4_150119850_150120001 | 0.25 |
Slc2a5 |
solute carrier family 2 (facilitated glucose transporter), member 5 |
642 |
0.38 |
| chr1_92834707_92834858 | 0.25 |
Gpc1 |
glypican 1 |
2813 |
0.15 |
| chr3_129589919_129590070 | 0.24 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
37614 |
0.13 |
| chr2_172511032_172511203 | 0.24 |
Gm14303 |
predicted gene 14303 |
1426 |
0.37 |
| chr2_94430791_94430942 | 0.24 |
Api5 |
apoptosis inhibitor 5 |
1122 |
0.47 |
| chr6_85919471_85919834 | 0.24 |
Gm18290 |
predicted gene, 18290 |
1766 |
0.16 |
| chr7_137068941_137069100 | 0.24 |
Gm45499 |
predicted gene 45499 |
98800 |
0.07 |
| chr16_87710092_87710297 | 0.24 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
11190 |
0.21 |
| chr1_131619346_131619811 | 0.23 |
Gm8532 |
predicted gene 8532 |
13391 |
0.14 |
| chr3_19225975_19226148 | 0.23 |
Gm16093 |
predicted gene 16093 |
1719 |
0.33 |
| chr7_19675959_19676128 | 0.23 |
Apoc2 |
apolipoprotein C-II |
821 |
0.36 |
| chr5_124236533_124236706 | 0.22 |
Gm42425 |
predicted gene 42425 |
1105 |
0.35 |
| chr8_45264437_45264634 | 0.22 |
F11 |
coagulation factor XI |
2504 |
0.25 |
| chr1_128440773_128440930 | 0.22 |
Dars |
aspartyl-tRNA synthetase |
23483 |
0.14 |
| chr8_121967787_121967995 | 0.22 |
Gm17709 |
predicted gene, 17709 |
5931 |
0.11 |
| chr1_194361449_194361616 | 0.22 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
138773 |
0.05 |
| chr9_55277532_55277801 | 0.22 |
Nrg4 |
neuregulin 4 |
5906 |
0.19 |
| chr8_95360199_95360374 | 0.21 |
Mmp15 |
matrix metallopeptidase 15 |
8018 |
0.13 |
| chr10_21328387_21328563 | 0.21 |
4930455C13Rik |
RIKEN cDNA 4930455C13 gene |
13582 |
0.14 |
| chr11_49233858_49234036 | 0.21 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
10244 |
0.12 |
| chr7_16615699_16615891 | 0.21 |
Gm44589 |
predicted gene 44589 |
556 |
0.43 |
| chr6_88737581_88737810 | 0.21 |
Gm44005 |
predicted gene, 44005 |
497 |
0.7 |
| chr3_10174352_10174597 | 0.21 |
Pmp2 |
peripheral myelin protein 2 |
9455 |
0.12 |
| chr18_50139098_50139320 | 0.20 |
Hsd17b4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
3160 |
0.23 |
| chr16_90282950_90283226 | 0.20 |
Scaf4 |
SR-related CTD-associated factor 4 |
1220 |
0.43 |
| chr18_44540279_44540430 | 0.20 |
Mcc |
mutated in colorectal cancers |
20838 |
0.24 |
| chr13_5714270_5714464 | 0.20 |
Gm35330 |
predicted gene, 35330 |
10075 |
0.27 |
| chr9_106235574_106235761 | 0.20 |
Alas1 |
aminolevulinic acid synthase 1 |
1417 |
0.25 |
| chr4_57250984_57251161 | 0.20 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
29018 |
0.17 |
| chr1_13312454_13312771 | 0.20 |
Ncoa2 |
nuclear receptor coactivator 2 |
12890 |
0.11 |
| chr2_11513206_11513376 | 0.19 |
Gm10851 |
predicted gene 10851 |
8415 |
0.14 |
| chr8_126309380_126309531 | 0.19 |
Slc35f3 |
solute carrier family 35, member F3 |
10897 |
0.27 |
| chr6_91424316_91424467 | 0.19 |
Gm4575 |
predicted gene 4575 |
4492 |
0.12 |
| chr5_124424178_124424509 | 0.19 |
Gm37415 |
predicted gene, 37415 |
354 |
0.67 |
| chr5_45857473_45857990 | 0.19 |
4930449I04Rik |
RIKEN cDNA 4930449I04 gene |
89 |
0.53 |
| chr9_72260390_72260691 | 0.19 |
Gm25163 |
predicted gene, 25163 |
3401 |
0.12 |
| chr7_140767390_140767578 | 0.18 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
3003 |
0.12 |
| chr5_144370961_144371328 | 0.18 |
Dmrt1i |
Dmrt1 interacting ncRNA |
12619 |
0.16 |
| chr9_13681735_13681905 | 0.18 |
Maml2 |
mastermind like transcriptional coactivator 2 |
19360 |
0.18 |
| chr15_77928675_77928831 | 0.18 |
Txn2 |
thioredoxin 2 |
215 |
0.92 |
| chr2_30545170_30545343 | 0.18 |
Gm14487 |
predicted gene 14487 |
10527 |
0.17 |
| chr4_124682416_124682588 | 0.17 |
Utp11 |
UTP11 small subunit processome component |
11032 |
0.09 |
| chr2_122075180_122075331 | 0.17 |
Spg11 |
SPG11, spatacsin vesicle trafficking associated |
43041 |
0.09 |
| chr16_78371572_78371756 | 0.16 |
Btg3 |
BTG anti-proliferation factor 3 |
1672 |
0.33 |
| chr16_7144291_7144454 | 0.16 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
49375 |
0.2 |
| chr2_119476960_119477373 | 0.16 |
Ino80 |
INO80 complex subunit |
447 |
0.8 |
| chr7_24850067_24850266 | 0.16 |
Gm18207 |
predicted gene, 18207 |
10780 |
0.08 |
| chr3_97642273_97642480 | 0.16 |
Fmo5 |
flavin containing monooxygenase 5 |
13493 |
0.12 |
| chr6_17463758_17464495 | 0.16 |
Met |
met proto-oncogene |
19 |
0.98 |
| chr2_27675051_27675434 | 0.15 |
Rxra |
retinoid X receptor alpha |
1198 |
0.58 |
| chr4_136675586_136676002 | 0.15 |
Ephb2 |
Eph receptor B2 |
14142 |
0.15 |
| chr14_61125286_61125469 | 0.15 |
Sacs |
sacsin |
13080 |
0.21 |
| chr2_152360517_152360697 | 0.15 |
Gm14165 |
predicted gene 14165 |
6992 |
0.09 |
| chr1_153691155_153691333 | 0.15 |
Gm29529 |
predicted gene 29529 |
1412 |
0.3 |
| chr5_127615097_127615417 | 0.15 |
Slc15a4 |
solute carrier family 15, member 4 |
1626 |
0.29 |
| chr5_122239684_122239867 | 0.15 |
Tctn1 |
tectonic family member 1 |
11903 |
0.12 |
| chrX_7422915_7423070 | 0.15 |
2010204K13Rik |
RIKEN cDNA 2010204K13 gene |
4 |
0.97 |
| chr8_70395705_70396002 | 0.15 |
Crtc1 |
CREB regulated transcription coactivator 1 |
6945 |
0.12 |
| chr19_43813317_43813611 | 0.15 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
2942 |
0.21 |
| chr5_114951424_114951834 | 0.15 |
2210016L21Rik |
RIKEN cDNA 2210016L21 gene |
5824 |
0.09 |
| chr4_46868313_46868486 | 0.14 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
8497 |
0.27 |
| chr12_73353785_73353952 | 0.14 |
Gm48653 |
predicted gene, 48653 |
1213 |
0.43 |
| chr11_51859695_51859870 | 0.14 |
Jade2 |
jade family PHD finger 2 |
2129 |
0.29 |
| chr5_146967227_146967378 | 0.14 |
Mtif3 |
mitochondrial translational initiation factor 3 |
3502 |
0.21 |
| chr4_122977007_122977265 | 0.14 |
4933421A08Rik |
RIKEN cDNA 4933421A08 gene |
15827 |
0.12 |
| chr15_55122695_55122962 | 0.14 |
Gm9920 |
predicted gene 9920 |
9151 |
0.15 |
| chrX_60008756_60008925 | 0.14 |
F9 |
coagulation factor IX |
9376 |
0.21 |
| chr4_123960615_123960766 | 0.14 |
Gm12902 |
predicted gene 12902 |
34456 |
0.11 |
| chr19_32524150_32524376 | 0.14 |
Gm36419 |
predicted gene, 36419 |
19085 |
0.15 |
| chr8_71700006_71700195 | 0.14 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
1689 |
0.18 |
| chr9_106447373_106447582 | 0.14 |
Abhd14b |
abhydrolase domain containing 14b |
18 |
0.49 |
| chr3_138256816_138256967 | 0.14 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
4100 |
0.14 |
| chr5_39219850_39220015 | 0.13 |
Gm40293 |
predicted gene, 40293 |
87152 |
0.08 |
| chr7_141064386_141064940 | 0.13 |
B4galnt4 |
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
169 |
0.88 |
| chr5_100430036_100430187 | 0.13 |
Gm42690 |
predicted gene 42690 |
385 |
0.59 |
| chrX_38423271_38423435 | 0.13 |
Lamp2 |
lysosomal-associated membrane protein 2 |
19797 |
0.15 |
| chr12_54405210_54405378 | 0.13 |
Gm46328 |
predicted gene, 46328 |
5118 |
0.16 |
| chr5_20704249_20704400 | 0.13 |
Magi2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
2655 |
0.3 |
| chr19_57368390_57368551 | 0.13 |
Fam160b1 |
family with sequence similarity 160, member B1 |
7440 |
0.16 |
| chr15_7154102_7154264 | 0.13 |
Lifr |
LIF receptor alpha |
170 |
0.97 |
| chr15_39111986_39112168 | 0.12 |
Slc25a32 |
solute carrier family 25, member 32 |
555 |
0.51 |
| chr11_116483917_116484077 | 0.12 |
Prpsap1 |
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
5677 |
0.11 |
| chr5_64114340_64114507 | 0.12 |
Pgm2 |
phosphoglucomutase 2 |
2322 |
0.2 |
| chr1_153697619_153697798 | 0.12 |
Gm29529 |
predicted gene 29529 |
5052 |
0.13 |
| chr10_42524541_42524692 | 0.12 |
Snx3 |
sorting nexin 3 |
22326 |
0.16 |
| chr2_91242516_91242810 | 0.12 |
Ddb2 |
damage specific DNA binding protein 2 |
5681 |
0.11 |
| chr18_68062225_68062529 | 0.12 |
Gm41764 |
predicted gene, 41764 |
55997 |
0.13 |
| chr11_65131461_65131612 | 0.12 |
1700086D15Rik |
RIKEN cDNA 1700086D15 gene |
28354 |
0.12 |
| chr10_44321957_44322108 | 0.12 |
Mir1929 |
microRNA 1929 |
37644 |
0.15 |
| chr15_76594336_76594507 | 0.12 |
Adck5 |
aarF domain containing kinase 5 |
128 |
0.89 |
| chr1_167746804_167746963 | 0.12 |
Lmx1a |
LIM homeobox transcription factor 1 alpha |
57326 |
0.15 |
| chr8_70372073_70372256 | 0.11 |
Comp |
cartilage oligomeric matrix protein |
1394 |
0.28 |
| chr11_120491297_120491754 | 0.11 |
Slc25a10 |
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
315 |
0.64 |
| chr2_120348240_120348405 | 0.11 |
Vps39 |
VPS39 HOPS complex subunit |
4115 |
0.19 |
| chr18_75502781_75502947 | 0.11 |
Gm10532 |
predicted gene 10532 |
11781 |
0.26 |
| chr12_105000981_105001188 | 0.11 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
2407 |
0.18 |
| chr5_123023127_123023430 | 0.11 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
7940 |
0.09 |
| chr6_114918998_114919299 | 0.11 |
Vgll4 |
vestigial like family member 4 |
2668 |
0.32 |
| chr6_100636259_100636566 | 0.11 |
Gm44108 |
predicted gene, 44108 |
9472 |
0.15 |
| chr8_3277578_3277729 | 0.11 |
Insr |
insulin receptor |
1898 |
0.34 |
| chr2_18543019_18543179 | 0.11 |
Gm13353 |
predicted gene 13353 |
25027 |
0.2 |
| chr13_80894659_80894898 | 0.11 |
Arrdc3 |
arrestin domain containing 3 |
4260 |
0.19 |
| chr16_33966526_33966697 | 0.11 |
Gm15829 |
predicted gene 15829 |
372 |
0.48 |
| chr4_59438657_59438836 | 0.11 |
Susd1 |
sushi domain containing 1 |
113 |
0.97 |
| chr16_52129443_52129606 | 0.11 |
Cblb |
Casitas B-lineage lymphoma b |
23002 |
0.21 |
| chr11_99105971_99106266 | 0.10 |
Tns4 |
tensin 4 |
16812 |
0.14 |
| chr11_101569721_101569872 | 0.10 |
Nbr1 |
NBR1, autophagy cargo receptor |
1109 |
0.3 |
| chr1_88411027_88411203 | 0.10 |
Spp2 |
secreted phosphoprotein 2 |
3709 |
0.2 |
| chr4_6908644_6908795 | 0.10 |
Tox |
thymocyte selection-associated high mobility group box |
81764 |
0.11 |
| chr1_168357153_168357454 | 0.10 |
Gm38381 |
predicted gene, 38381 |
6495 |
0.24 |
| chr1_170815209_170815368 | 0.10 |
Gm25235 |
predicted gene, 25235 |
17583 |
0.12 |
| chr17_34898174_34898489 | 0.10 |
C2 |
complement component 2 (within H-2S) |
66 |
0.57 |
| chr7_114461865_114462041 | 0.10 |
Pde3b |
phosphodiesterase 3B, cGMP-inhibited |
44225 |
0.15 |
| chr8_62184095_62184246 | 0.10 |
Gm2961 |
predicted gene 2961 |
10042 |
0.27 |
| chr5_111509539_111510088 | 0.10 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
71609 |
0.09 |
| chr12_31950204_31950930 | 0.10 |
Hbp1 |
high mobility group box transcription factor 1 |
32 |
0.98 |
| chr1_52499794_52500020 | 0.10 |
Nab1 |
Ngfi-A binding protein 1 |
393 |
0.8 |
| chr6_99145207_99145385 | 0.10 |
Foxp1 |
forkhead box P1 |
17722 |
0.26 |
| chr7_81588668_81588888 | 0.10 |
Gm45698 |
predicted gene 45698 |
2116 |
0.19 |
| chr2_35211769_35211922 | 0.09 |
Rab14 |
RAB14, member RAS oncogene family |
10725 |
0.14 |
| chr6_95163817_95164232 | 0.09 |
Kbtbd8 |
kelch repeat and BTB (POZ) domain containing 8 |
46040 |
0.12 |
| chr3_35892773_35892978 | 0.09 |
Dcun1d1 |
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
16970 |
0.12 |
| chr8_120046377_120046540 | 0.09 |
Gm15684 |
predicted gene 15684 |
1171 |
0.4 |
| chr8_13055180_13055331 | 0.09 |
Proz |
protein Z, vitamin K-dependent plasma glycoprotein |
5659 |
0.1 |
| chr6_116015696_116015857 | 0.09 |
Plxnd1 |
plexin D1 |
20771 |
0.14 |
| chr6_108662142_108662337 | 0.09 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
807 |
0.49 |
| chr4_141452237_141452709 | 0.09 |
Zbtb17 |
zinc finger and BTB domain containing 17 |
7767 |
0.11 |
| chr11_101700898_101701098 | 0.09 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
31921 |
0.08 |
| chr2_3424682_3424836 | 0.09 |
Dclre1c |
DNA cross-link repair 1C |
557 |
0.63 |
| chr18_61373998_61374172 | 0.08 |
Gm25301 |
predicted gene, 25301 |
23728 |
0.12 |
| chr7_49351868_49352264 | 0.08 |
Gm44913 |
predicted gene 44913 |
5972 |
0.24 |
| chr11_55468108_55468290 | 0.08 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1486 |
0.25 |
| chr11_82852598_82853205 | 0.08 |
Rffl |
ring finger and FYVE like domain containing protein |
6809 |
0.12 |
| chr2_48442189_48442365 | 0.08 |
Gm13481 |
predicted gene 13481 |
14968 |
0.23 |
| chr9_54844002_54844168 | 0.08 |
Rab7-ps1 |
RAB7, member RAS oncogene family, pseudogene 1 |
18803 |
0.15 |
| chr8_40907855_40908028 | 0.08 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
9503 |
0.16 |
| chr3_32399514_32399821 | 0.08 |
Pik3ca |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
1996 |
0.29 |
| chr9_18293076_18293227 | 0.08 |
Chordc1 |
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
763 |
0.59 |
| chr19_40471969_40472156 | 0.08 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
15493 |
0.2 |
| chr6_144311505_144311701 | 0.08 |
Sox5 |
SRY (sex determining region Y)-box 5 |
102035 |
0.08 |
| chr2_101831932_101832083 | 0.08 |
Prr5l |
proline rich 5 like |
6973 |
0.23 |
| chr11_88181207_88181562 | 0.08 |
Cuedc1 |
CUE domain containing 1 |
919 |
0.56 |
| chr18_33780213_33780407 | 0.08 |
Epb41l4aos |
erythrocyte membrane protein band 4.1 like 4a, opposite strand |
14582 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.7 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.0 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |