Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
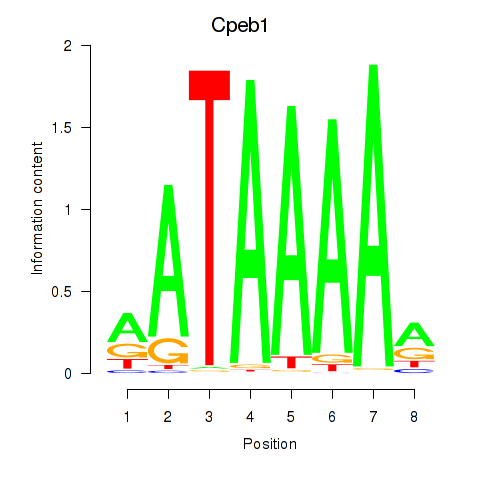
Results for Cpeb1
Z-value: 2.10
Transcription factors associated with Cpeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cpeb1
|
ENSMUSG00000025586.10 | cytoplasmic polyadenylation element binding protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_81453890_81454046 | Cpeb1 | 707 | 0.508561 | 0.76 | 7.7e-02 | Click! |
| chr7_81368113_81368268 | Cpeb1 | 4251 | 0.187287 | 0.74 | 9.0e-02 | Click! |
| chr7_81455393_81455591 | Cpeb1 | 27 | 0.857071 | 0.74 | 9.1e-02 | Click! |
| chr7_81362482_81362652 | Cpeb1 | 6572 | 0.167095 | 0.57 | 2.4e-01 | Click! |
| chr7_81379538_81379821 | Cpeb1 | 7194 | 0.165597 | 0.41 | 4.2e-01 | Click! |
Activity of the Cpeb1 motif across conditions
Conditions sorted by the z-value of the Cpeb1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_69139928_69140109 | 0.97 |
Gm1647 |
predicted gene 1647 |
8785 |
0.14 |
| chr14_70338039_70338256 | 0.88 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
3949 |
0.15 |
| chr5_135741186_135741337 | 0.80 |
Tmem120a |
transmembrane protein 120A |
1138 |
0.32 |
| chr16_23456512_23456863 | 0.75 |
Rtp1 |
receptor transporter protein 1 |
27554 |
0.13 |
| chr16_13258895_13259057 | 0.71 |
Mrtfb |
myocardin related transcription factor B |
2470 |
0.38 |
| chr14_48714246_48714540 | 0.63 |
Gm49153 |
predicted gene, 49153 |
10209 |
0.1 |
| chr2_68873804_68874167 | 0.61 |
Cers6 |
ceramide synthase 6 |
12399 |
0.14 |
| chr2_69143255_69143420 | 0.60 |
Nostrin |
nitric oxide synthase trafficker |
7537 |
0.21 |
| chr5_146114373_146114524 | 0.59 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
35181 |
0.09 |
| chr3_27909545_27909755 | 0.59 |
Tmem212 |
transmembrane protein 212 |
13282 |
0.21 |
| chr13_94250958_94251141 | 0.57 |
Scamp1 |
secretory carrier membrane protein 1 |
4491 |
0.2 |
| chr15_93916844_93917358 | 0.54 |
9430014N10Rik |
RIKEN cDNA 9430014N10 gene |
10010 |
0.25 |
| chr5_8962552_8962757 | 0.54 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
4102 |
0.15 |
| chr2_169997535_169997686 | 0.53 |
AY702102 |
cDNA sequence AY702102 |
34423 |
0.21 |
| chr10_19443595_19443775 | 0.52 |
Gm33104 |
predicted gene, 33104 |
43030 |
0.15 |
| chr18_70622479_70622643 | 0.52 |
Mbd2 |
methyl-CpG binding domain protein 2 |
4788 |
0.23 |
| chr1_84877752_84878205 | 0.52 |
Fbxo36 |
F-box protein 36 |
8827 |
0.16 |
| chr9_101036260_101036546 | 0.51 |
Pccb |
propionyl Coenzyme A carboxylase, beta polypeptide |
1505 |
0.33 |
| chr17_84074449_84074742 | 0.51 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
4065 |
0.18 |
| chr15_101320859_101321043 | 0.51 |
Gm35853 |
predicted gene, 35853 |
1937 |
0.18 |
| chr6_15232040_15232214 | 0.50 |
Foxp2 |
forkhead box P2 |
22156 |
0.28 |
| chr12_117720585_117720755 | 0.50 |
Gm18955 |
predicted gene, 18955 |
3302 |
0.28 |
| chr2_155538152_155538306 | 0.50 |
Mipep-ps |
mitochondrial intermediate peptidase, pseudogene |
4418 |
0.1 |
| chr3_41569165_41569423 | 0.50 |
Jade1 |
jade family PHD finger 1 |
4374 |
0.18 |
| chr9_73050700_73050869 | 0.49 |
Rab27a |
RAB27A, member RAS oncogene family |
5800 |
0.09 |
| chr14_37027574_37027733 | 0.48 |
Gm47885 |
predicted gene, 47885 |
3119 |
0.2 |
| chr2_155527626_155527777 | 0.48 |
Ggt7 |
gamma-glutamyltransferase 7 |
9464 |
0.09 |
| chr8_105994175_105994329 | 0.48 |
Dpep2 |
dipeptidase 2 |
2142 |
0.12 |
| chr10_116221046_116221335 | 0.48 |
Ptprr |
protein tyrosine phosphatase, receptor type, R |
24794 |
0.18 |
| chr17_29496086_29496540 | 0.48 |
Pim1 |
proviral integration site 1 |
2906 |
0.15 |
| chr14_14121099_14121351 | 0.48 |
Psmd6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
241 |
0.9 |
| chr5_145975985_145976140 | 0.48 |
Gm43115 |
predicted gene 43115 |
7652 |
0.13 |
| chr7_123193136_123193287 | 0.47 |
Tnrc6a |
trinucleotide repeat containing 6a |
13290 |
0.2 |
| chr7_46856262_46856448 | 0.46 |
Ldhc |
lactate dehydrogenase C |
4848 |
0.11 |
| chr12_102451508_102451858 | 0.46 |
Gm30198 |
predicted gene, 30198 |
4355 |
0.19 |
| chr15_90992970_90993407 | 0.46 |
Kif21a |
kinesin family member 21A |
26786 |
0.16 |
| chr18_47313549_47313714 | 0.46 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
19695 |
0.21 |
| chr17_63076780_63076958 | 0.45 |
Gm25348 |
predicted gene, 25348 |
20131 |
0.27 |
| chr3_85819269_85819420 | 0.45 |
Fam160a1 |
family with sequence similarity 160, member A1 |
2053 |
0.31 |
| chr13_18736718_18737047 | 0.45 |
Vps41 |
VPS41 HOPS complex subunit |
19592 |
0.25 |
| chr18_4683358_4683671 | 0.45 |
Jcad |
junctional cadherin 5 associated |
34446 |
0.18 |
| chr4_123576766_123577231 | 0.45 |
Macf1 |
microtubule-actin crosslinking factor 1 |
3938 |
0.22 |
| chr11_100322740_100322903 | 0.45 |
Eif1 |
eukaryotic translation initiation factor 1 |
1937 |
0.15 |
| chr1_165348964_165349116 | 0.43 |
Dcaf6 |
DDB1 and CUL4 associated factor 6 |
2958 |
0.22 |
| chr8_125405126_125405301 | 0.43 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
87497 |
0.09 |
| chr6_145355079_145355230 | 0.43 |
Gm23498 |
predicted gene, 23498 |
11704 |
0.13 |
| chr4_47315471_47315622 | 0.43 |
Col15a1 |
collagen, type XV, alpha 1 |
11824 |
0.22 |
| chr4_150105593_150105744 | 0.43 |
AL606971.1 |
solute carrier family 2 pseudogene |
6757 |
0.11 |
| chr19_3831408_3831611 | 0.42 |
Gm19209 |
predicted gene, 19209 |
9120 |
0.1 |
| chr15_55866367_55866558 | 0.42 |
Sntb1 |
syntrophin, basic 1 |
39848 |
0.15 |
| chr9_105809298_105809523 | 0.42 |
Col6a6 |
collagen, type VI, alpha 6 |
238 |
0.94 |
| chr12_85825843_85826036 | 0.42 |
Ttll5 |
tubulin tyrosine ligase-like family, member 5 |
545 |
0.68 |
| chr8_128462997_128463148 | 0.42 |
Nrp1 |
neuropilin 1 |
103675 |
0.07 |
| chr13_111431183_111431339 | 0.41 |
Gpbp1 |
GC-rich promoter binding protein 1 |
22121 |
0.17 |
| chr5_145980008_145980370 | 0.41 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
11454 |
0.12 |
| chr5_118301077_118301242 | 0.41 |
Gm25076 |
predicted gene, 25076 |
34710 |
0.14 |
| chr13_52976319_52976492 | 0.41 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
4668 |
0.2 |
| chr12_40508704_40509084 | 0.41 |
Dock4 |
dedicator of cytokinesis 4 |
62558 |
0.12 |
| chr12_118875203_118875358 | 0.41 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
15479 |
0.21 |
| chrX_164035471_164035631 | 0.40 |
Car5b |
carbonic anhydrase 5b, mitochondrial |
7554 |
0.2 |
| chr18_7847563_7847725 | 0.40 |
Gm50027 |
predicted gene, 50027 |
2709 |
0.23 |
| chr19_47447791_47448053 | 0.40 |
Sh3pxd2a |
SH3 and PX domains 2A |
16325 |
0.19 |
| chr8_11055813_11056042 | 0.40 |
9530052E02Rik |
RIKEN cDNA 9530052E02 gene |
6349 |
0.16 |
| chr9_12511948_12512243 | 0.40 |
Gm25365 |
predicted gene, 25365 |
268070 |
0.02 |
| chr6_87428535_87428790 | 0.40 |
Bmp10 |
bone morphogenetic protein 10 |
332 |
0.85 |
| chr4_81591457_81591882 | 0.39 |
Gm11411 |
predicted gene 11411 |
104772 |
0.07 |
| chr18_84600368_84600897 | 0.39 |
Zfp407 |
zinc finger protein 407 |
10907 |
0.16 |
| chr3_69778568_69778734 | 0.39 |
Nmd3 |
NMD3 ribosome export adaptor |
32219 |
0.16 |
| chr6_100629048_100629393 | 0.39 |
Gm44108 |
predicted gene, 44108 |
2280 |
0.25 |
| chr5_87139047_87139701 | 0.39 |
Ugt2b5 |
UDP glucuronosyltransferase 2 family, polypeptide B5 |
944 |
0.44 |
| chr1_167649230_167649384 | 0.39 |
Rxrg |
retinoid X receptor gamma |
30998 |
0.19 |
| chr8_11149308_11149679 | 0.39 |
Gm44717 |
predicted gene 44717 |
247 |
0.91 |
| chrX_60405283_60405665 | 0.39 |
Atp11c |
ATPase, class VI, type 11C |
1493 |
0.41 |
| chr2_29731870_29732021 | 0.39 |
Rapgef1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
616 |
0.61 |
| chr2_132130983_132131187 | 0.39 |
Gm14051 |
predicted gene 14051 |
13448 |
0.16 |
| chr3_88685605_88685767 | 0.39 |
Khdc4 |
KH domain containing 4, pre-mRNA splicing factor |
117 |
0.86 |
| chr2_12189127_12189278 | 0.39 |
Itga8 |
integrin alpha 8 |
4110 |
0.28 |
| chrX_167880328_167880485 | 0.39 |
Frmpd4 |
FERM and PDZ domain containing 4 |
17152 |
0.21 |
| chr10_127213325_127213485 | 0.39 |
Pip4k2c |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
1820 |
0.17 |
| chr18_5945440_5945591 | 0.39 |
Gm34804 |
predicted gene, 34804 |
332 |
0.92 |
| chr7_89423464_89423896 | 0.38 |
AI314278 |
expressed sequence AI314278 |
2626 |
0.22 |
| chr2_90650072_90650249 | 0.38 |
Nup160 |
nucleoporin 160 |
27055 |
0.17 |
| chr6_71824307_71824486 | 0.38 |
Mrpl35 |
mitochondrial ribosomal protein L35 |
586 |
0.6 |
| chr18_15408685_15408836 | 0.38 |
Aqp4 |
aquaporin 4 |
672 |
0.72 |
| chr10_75407573_75407724 | 0.38 |
Upb1 |
ureidopropionase, beta |
690 |
0.67 |
| chr11_6344574_6344748 | 0.38 |
Zmiz2 |
zinc finger, MIZ-type containing 2 |
44413 |
0.07 |
| chr10_84055805_84055963 | 0.37 |
Gm37908 |
predicted gene, 37908 |
5874 |
0.21 |
| chr11_108936283_108936686 | 0.37 |
Axin2 |
axin 2 |
2390 |
0.3 |
| chr11_69110217_69110963 | 0.36 |
Hes7 |
hes family bHLH transcription factor 7 |
9814 |
0.07 |
| chr16_95682151_95682417 | 0.36 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
19791 |
0.2 |
| chr16_80273340_80273491 | 0.36 |
Gm23083 |
predicted gene, 23083 |
136063 |
0.05 |
| chr13_64349576_64349754 | 0.36 |
Gm47123 |
predicted gene, 47123 |
11387 |
0.1 |
| chr15_10183843_10184024 | 0.36 |
Prlr |
prolactin receptor |
6152 |
0.31 |
| chr2_146533260_146533500 | 0.36 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
9551 |
0.24 |
| chr6_129369459_129369622 | 0.36 |
Clec12a |
C-type lectin domain family 12, member a |
12738 |
0.11 |
| chr1_36537159_36537318 | 0.36 |
Ankrd23 |
ankyrin repeat domain 23 |
355 |
0.71 |
| chr17_80051656_80051839 | 0.36 |
Gpx4-ps1 |
glutathione peroxidase 4, pseudogene 1 |
2279 |
0.23 |
| chr11_45255693_45255855 | 0.36 |
Gm12161 |
predicted gene 12161 |
25180 |
0.18 |
| chr7_96960523_96960692 | 0.36 |
C230038L03Rik |
RIKEN cDNA C230038L03 gene |
8721 |
0.18 |
| chr3_106493950_106494113 | 0.36 |
Dennd2d |
DENN/MADD domain containing 2D |
856 |
0.5 |
| chr13_92469480_92469792 | 0.36 |
Fam151b |
family with sequence similarity 151, member B |
14377 |
0.17 |
| chr11_50188709_50189126 | 0.36 |
Mrnip |
MRN complex interacting protein |
14010 |
0.1 |
| chr2_80600719_80600876 | 0.35 |
Gm13688 |
predicted gene 13688 |
1936 |
0.21 |
| chr4_108166700_108166861 | 0.35 |
Echdc2 |
enoyl Coenzyme A hydratase domain containing 2 |
1262 |
0.4 |
| chr16_8711657_8711808 | 0.35 |
Usp7 |
ubiquitin specific peptidase 7 |
4805 |
0.16 |
| chr10_80441787_80441955 | 0.35 |
Tcf3 |
transcription factor 3 |
8224 |
0.09 |
| chr7_25837976_25838184 | 0.35 |
Gm4613 |
predicted gene 4613 |
9203 |
0.08 |
| chr5_139343215_139343377 | 0.35 |
Cox19 |
cytochrome c oxidase assembly protein 19 |
1829 |
0.23 |
| chr2_155059266_155059435 | 0.35 |
a |
nonagouti |
11735 |
0.13 |
| chr11_50189181_50189338 | 0.35 |
Sqstm1 |
sequestosome 1 |
13695 |
0.1 |
| chr5_118332124_118332291 | 0.35 |
Gm28563 |
predicted gene 28563 |
65123 |
0.09 |
| chr5_45513758_45514082 | 0.35 |
Lap3 |
leucine aminopeptidase 3 |
4711 |
0.13 |
| chr1_88764323_88764494 | 0.35 |
Platr5 |
pluripotency associated transcript 5 |
9454 |
0.2 |
| chr2_160990763_160990931 | 0.34 |
Chd6 |
chromodomain helicase DNA binding protein 6 |
5801 |
0.18 |
| chr18_43980831_43980982 | 0.34 |
Spink5 |
serine peptidase inhibitor, Kazal type 5 |
17671 |
0.17 |
| chr9_118763193_118763462 | 0.34 |
Itga9 |
integrin alpha 9 |
43957 |
0.15 |
| chr19_6264376_6264541 | 0.34 |
Mir194-2 |
microRNA 194-2 |
185 |
0.67 |
| chr3_50989485_50989700 | 0.34 |
Gm37209 |
predicted gene, 37209 |
10781 |
0.22 |
| chr4_126091997_126092189 | 0.34 |
Oscp1 |
organic solute carrier partner 1 |
4414 |
0.13 |
| chr5_118675758_118675934 | 0.34 |
Gm43052 |
predicted gene 43052 |
11772 |
0.16 |
| chr6_26500672_26500834 | 0.34 |
Gm24691 |
predicted gene, 24691 |
40323 |
0.23 |
| chr12_101066448_101066619 | 0.34 |
Ppp4r3a |
protein phosphatase 4 regulatory subunit 3A |
15127 |
0.11 |
| chr3_77937446_77937597 | 0.34 |
Gm18719 |
predicted gene, 18719 |
23863 |
0.22 |
| chr8_129135025_129135198 | 0.34 |
Ccdc7b |
coiled-coil domain containing 7B |
1409 |
0.38 |
| chr18_74418143_74418457 | 0.34 |
Gm50168 |
predicted gene, 50168 |
2505 |
0.25 |
| chr16_22698076_22698372 | 0.34 |
Gm8118 |
predicted gene 8118 |
12030 |
0.18 |
| chr10_94221527_94221700 | 0.34 |
Ndufa12 |
NADH:ubiquinone oxidoreductase subunit A12 |
22576 |
0.15 |
| chr4_97757677_97758345 | 0.34 |
Gm12676 |
predicted gene 12676 |
622 |
0.75 |
| chr14_117680704_117680890 | 0.34 |
Mir6239 |
microRNA 6239 |
273050 |
0.01 |
| chr6_91752974_91753164 | 0.34 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
2885 |
0.21 |
| chr17_32257244_32257401 | 0.34 |
Brd4 |
bromodomain containing 4 |
15043 |
0.12 |
| chr6_71205289_71205700 | 0.33 |
Fabp1 |
fatty acid binding protein 1, liver |
5667 |
0.13 |
| chr8_119429638_119429810 | 0.33 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
4400 |
0.17 |
| chr2_169634592_169634925 | 0.33 |
Tshz2 |
teashirt zinc finger family member 2 |
1082 |
0.55 |
| chr15_103280956_103281148 | 0.33 |
Mir148b |
microRNA 148b |
4073 |
0.11 |
| chr19_38223407_38224029 | 0.33 |
Fra10ac1 |
FRA10AC1 homolog (human) |
420 |
0.83 |
| chr3_28301524_28301675 | 0.33 |
Gm37537 |
predicted gene, 37537 |
22758 |
0.19 |
| chr15_9076605_9076777 | 0.33 |
Nadk2 |
NAD kinase 2, mitochondrial |
1414 |
0.47 |
| chr5_147751844_147752098 | 0.33 |
Gm43156 |
predicted gene 43156 |
6455 |
0.21 |
| chr2_170176271_170176422 | 0.33 |
Zfp217 |
zinc finger protein 217 |
28243 |
0.22 |
| chr2_132654710_132655013 | 0.33 |
Rpl23a-ps4 |
ribosomal protein L23A, pseudogene 4 |
7381 |
0.12 |
| chr10_8086797_8087175 | 0.33 |
Gm48614 |
predicted gene, 48614 |
65694 |
0.11 |
| chr13_9761902_9762194 | 0.33 |
Zmynd11 |
zinc finger, MYND domain containing 11 |
2302 |
0.24 |
| chr3_148429134_148429295 | 0.32 |
Gm43576 |
predicted gene 43576 |
25727 |
0.24 |
| chr5_134170212_134170373 | 0.32 |
Rcc1l |
reculator of chromosome condensation 1 like |
917 |
0.46 |
| chr16_58501861_58502070 | 0.32 |
St3gal6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
2624 |
0.27 |
| chr11_70713419_70713657 | 0.32 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr1_134357912_134358070 | 0.32 |
Tmem183a |
transmembrane protein 183A |
3699 |
0.15 |
| chr5_134583600_134583751 | 0.32 |
Rfc2 |
replication factor C (activator 1) 2 |
918 |
0.4 |
| chrX_38473317_38473492 | 0.32 |
Gm7598 |
predicted gene 7598 |
3057 |
0.24 |
| chr10_123843693_123843994 | 0.32 |
Gm18510 |
predicted gene, 18510 |
48055 |
0.2 |
| chr7_88291376_88291631 | 0.32 |
Ctsc |
cathepsin C |
5349 |
0.23 |
| chr12_58323949_58324220 | 0.32 |
Clec14a |
C-type lectin domain family 14, member a |
54794 |
0.16 |
| chr4_80004164_80004316 | 0.32 |
Gm11410 |
predicted gene 11410 |
251 |
0.57 |
| chr11_101378888_101379088 | 0.32 |
G6pc |
glucose-6-phosphatase, catalytic |
11427 |
0.06 |
| chr14_51106687_51106910 | 0.32 |
Eddm3b |
epididymal protein 3B |
7628 |
0.08 |
| chr1_136955166_136955491 | 0.32 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
1698 |
0.42 |
| chr3_129549658_129549814 | 0.32 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
2644 |
0.24 |
| chr2_145680265_145680416 | 0.32 |
Rin2 |
Ras and Rab interactor 2 |
4973 |
0.24 |
| chr19_21897509_21897685 | 0.32 |
Ldhb-ps |
lactate dehydrogenase B, pseudogene |
39783 |
0.14 |
| chr2_3113922_3114235 | 0.32 |
Fam171a1 |
family with sequence similarity 171, member A1 |
146 |
0.97 |
| chr17_30010480_30010631 | 0.32 |
Zfand3 |
zinc finger, AN1-type domain 3 |
2758 |
0.19 |
| chr7_140722373_140722532 | 0.32 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
1170 |
0.3 |
| chr18_10461260_10461439 | 0.31 |
Greb1l |
growth regulation by estrogen in breast cancer-like |
3124 |
0.25 |
| chr4_59761101_59761273 | 0.31 |
Inip |
INTS3 and NABP interacting protein |
22621 |
0.16 |
| chr7_72345699_72346292 | 0.31 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
39387 |
0.21 |
| chr8_117334963_117335140 | 0.31 |
Cmip |
c-Maf inducing protein |
14119 |
0.25 |
| chr5_91200748_91201014 | 0.31 |
Gm23092 |
predicted gene, 23092 |
17123 |
0.2 |
| chr14_45418421_45418590 | 0.31 |
Gm25353 |
predicted gene, 25353 |
13545 |
0.12 |
| chr1_58111776_58112051 | 0.31 |
Aox3 |
aldehyde oxidase 3 |
1217 |
0.49 |
| chr2_15096629_15097014 | 0.31 |
Arl5b |
ADP-ribosylation factor-like 5B |
21822 |
0.14 |
| chr11_86760028_86760187 | 0.31 |
Cltc |
clathrin, heavy polypeptide (Hc) |
2542 |
0.29 |
| chr3_148109608_148109780 | 0.31 |
Gm43574 |
predicted gene 43574 |
20550 |
0.29 |
| chr12_72637596_72637773 | 0.31 |
Gm4756 |
predicted gene 4756 |
8750 |
0.17 |
| chr4_105731258_105731470 | 0.31 |
Gm12728 |
predicted gene 12728 |
58505 |
0.15 |
| chr1_139548791_139549059 | 0.31 |
Cfhr1 |
complement factor H-related 1 |
4674 |
0.22 |
| chr14_114062877_114063028 | 0.31 |
Gm18369 |
predicted gene, 18369 |
109491 |
0.07 |
| chr4_155566693_155567032 | 0.31 |
Nadk |
NAD kinase |
2790 |
0.15 |
| chr5_116435813_116435964 | 0.31 |
Srrm4os |
serine/arginine repetitive matrix 4, opposite strand |
2832 |
0.18 |
| chr13_112344459_112344615 | 0.31 |
Gm48837 |
predicted gene, 48837 |
23308 |
0.14 |
| chr16_24223745_24224269 | 0.31 |
Gm31814 |
predicted gene, 31814 |
7483 |
0.23 |
| chr6_21852425_21852684 | 0.31 |
Tspan12 |
tetraspanin 12 |
39 |
0.62 |
| chr6_136518661_136518819 | 0.31 |
Atf7ip |
activating transcription factor 7 interacting protein |
62 |
0.78 |
| chr5_30168796_30168956 | 0.31 |
Mir1960 |
microRNA 1960 |
1863 |
0.23 |
| chr2_124741001_124741334 | 0.31 |
Gm13994 |
predicted gene 13994 |
32512 |
0.23 |
| chr9_89218115_89218458 | 0.31 |
Mthfs |
5, 10-methenyltetrahydrofolate synthetase |
168 |
0.93 |
| chr3_131371095_131371256 | 0.30 |
Gm43116 |
predicted gene 43116 |
10619 |
0.19 |
| chr6_48666335_48666494 | 0.30 |
Gm44262 |
predicted gene, 44262 |
554 |
0.52 |
| chr3_36475062_36475233 | 0.30 |
Mir7009 |
microRNA 7009 |
377 |
0.48 |
| chr13_35015805_35015976 | 0.30 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
11204 |
0.12 |
| chr6_88193601_88193754 | 0.30 |
Gata2 |
GATA binding protein 2 |
214 |
0.9 |
| chr6_72028118_72028756 | 0.30 |
9130221F21Rik |
RIKEN cDNA 9130221F21 gene |
14091 |
0.13 |
| chr7_25987036_25987409 | 0.30 |
Gm45225 |
predicted gene 45225 |
5720 |
0.14 |
| chr2_42039219_42039481 | 0.30 |
Gm13461 |
predicted gene 13461 |
50521 |
0.18 |
| chr6_137145657_137145998 | 0.30 |
4930480K02Rik |
RIKEN cDNA 4930480K02 gene |
1102 |
0.54 |
| chr2_73919805_73919982 | 0.30 |
Atp5g3 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
8567 |
0.2 |
| chr19_47860475_47860640 | 0.30 |
Gsto1 |
glutathione S-transferase omega 1 |
2438 |
0.22 |
| chr19_39286940_39287122 | 0.30 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
43 |
0.98 |
| chr9_124143748_124143915 | 0.30 |
Ccr5 |
chemokine (C-C motif) receptor 5 |
15083 |
0.2 |
| chr12_101875225_101875382 | 0.30 |
Trip11 |
thyroid hormone receptor interactor 11 |
8304 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 0.6 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.3 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 0.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.7 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 0.2 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.2 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.3 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.3 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.2 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.1 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.2 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0003097 | renal water transport(GO:0003097) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.3 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0097476 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.2 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.0 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.0 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0014745 | negative regulation of muscle adaptation(GO:0014745) |
| 0.0 | 0.0 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.0 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.0 | 0.1 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.3 | GO:0009813 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.0 | GO:0032762 | mast cell cytokine production(GO:0032762) |
| 0.0 | 0.0 | GO:1900221 | regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.0 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0001802 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:1904705 | regulation of vascular smooth muscle cell proliferation(GO:1904705) vascular smooth muscle cell proliferation(GO:1990874) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.4 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.4 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0034821 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0034812 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |