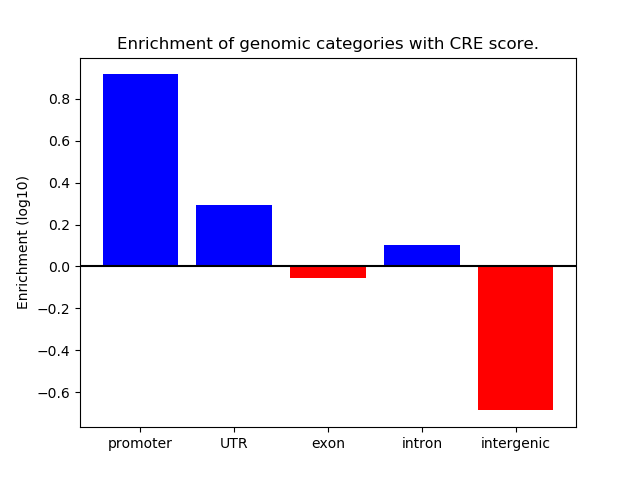
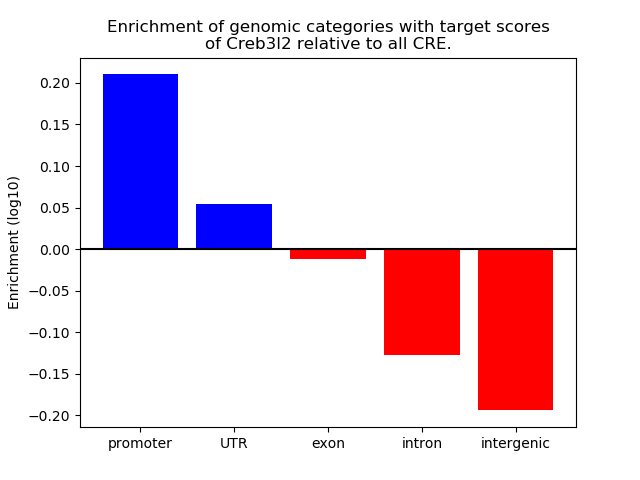
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
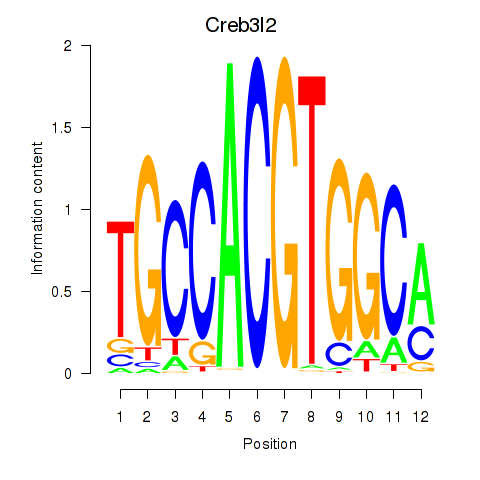
Results for Creb3l2
Z-value: 2.46
Transcription factors associated with Creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3l2
|
ENSMUSG00000038648.5 | cAMP responsive element binding protein 3-like 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_37480097_37480265 | Creb3l2 | 38035 | 0.166799 | -0.90 | 1.5e-02 | Click! |
| chr6_37431530_37431865 | Creb3l2 | 10449 | 0.271981 | -0.88 | 2.0e-02 | Click! |
| chr6_37441336_37441654 | Creb3l2 | 651 | 0.801531 | 0.84 | 3.7e-02 | Click! |
| chr6_37426047_37426198 | Creb3l2 | 16024 | 0.258549 | -0.79 | 6.2e-02 | Click! |
| chr6_37448065_37448224 | Creb3l2 | 5998 | 0.285415 | 0.78 | 6.5e-02 | Click! |
Activity of the Creb3l2 motif across conditions
Conditions sorted by the z-value of the Creb3l2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
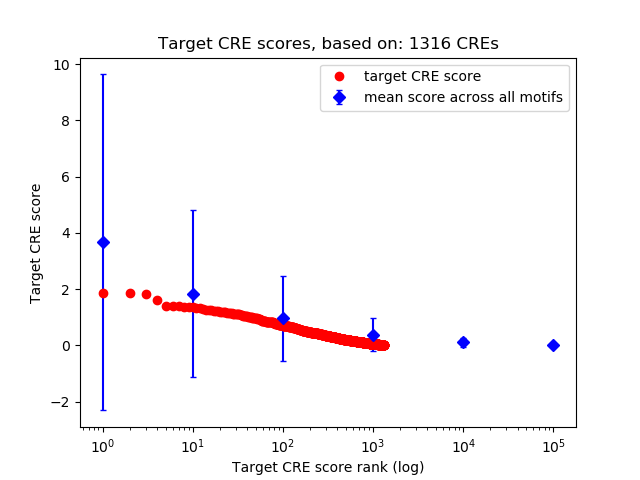
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_112103112_112103298 | 1.87 |
Mir128-2 |
microRNA 128-2 |
15506 |
0.25 |
| chr4_138981088_138981239 | 1.85 |
Tmco4 |
transmembrane and coiled-coil domains 4 |
8254 |
0.16 |
| chr19_46056737_46057343 | 1.81 |
Pprc1 |
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
176 |
0.92 |
| chr9_60982764_60983106 | 1.59 |
Gm5122 |
predicted gene 5122 |
30546 |
0.14 |
| chr2_163515893_163516044 | 1.41 |
Gm25881 |
predicted gene, 25881 |
1452 |
0.28 |
| chr10_93077177_93077387 | 1.40 |
Cfap54 |
cilia and flagella associated protein 54 |
4314 |
0.23 |
| chr15_62044174_62044346 | 1.40 |
Pvt1 |
Pvt1 oncogene |
5003 |
0.28 |
| chr4_117120836_117121181 | 1.38 |
Btbd19 |
BTB (POZ) domain containing 19 |
257 |
0.77 |
| chr2_52298505_52298656 | 1.37 |
Gm22095 |
predicted gene, 22095 |
15503 |
0.19 |
| chr18_60848442_60848618 | 1.36 |
Tcof1 |
treacle ribosome biogenesis factor 1 |
84 |
0.96 |
| chr4_148591704_148592085 | 1.32 |
Srm |
spermidine synthase |
24 |
0.95 |
| chr9_106234042_106234256 | 1.31 |
Alas1 |
aminolevulinic acid synthase 1 |
2935 |
0.14 |
| chr15_97035420_97035580 | 1.30 |
Slc38a4 |
solute carrier family 38, member 4 |
4289 |
0.32 |
| chr9_21367416_21367785 | 1.27 |
Ilf3 |
interleukin enhancer binding factor 3 |
271 |
0.81 |
| chr10_77902990_77903176 | 1.25 |
Lrrc3 |
leucine rich repeat containing 3 |
547 |
0.54 |
| chr5_64298676_64298855 | 1.25 |
Tbc1d1 |
TBC1 domain family, member 1 |
235 |
0.91 |
| chr1_180223322_180223519 | 1.21 |
Psen2 |
presenilin 2 |
6148 |
0.15 |
| chr2_174343758_174344168 | 1.21 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
349 |
0.83 |
| chr4_142337823_142337991 | 1.20 |
Gm13052 |
predicted gene 13052 |
8568 |
0.28 |
| chr10_59295146_59295479 | 1.20 |
P4ha1 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
27984 |
0.16 |
| chr5_31094198_31094360 | 1.20 |
Slc30a3 |
solute carrier family 30 (zinc transporter), member 3 |
522 |
0.44 |
| chr9_83834067_83834712 | 1.20 |
Ttk |
Ttk protein kinase |
300 |
0.91 |
| chr2_78656286_78656827 | 1.19 |
Gm14463 |
predicted gene 14463 |
873 |
0.71 |
| chr8_93179883_93180034 | 1.16 |
Ces1d |
carboxylesterase 1D |
4669 |
0.15 |
| chr7_142533086_142533550 | 1.16 |
Mrpl23 |
mitochondrial ribosomal protein L23 |
156 |
0.91 |
| chr10_127063341_127063519 | 1.15 |
Cdk4 |
cyclin-dependent kinase 4 |
104 |
0.9 |
| chr11_113148197_113148382 | 1.14 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
24788 |
0.24 |
| chr8_14911524_14911687 | 1.13 |
Arhgef10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
58 |
0.97 |
| chr3_52622106_52622284 | 1.12 |
Gm10293 |
predicted pseudogene 10293 |
9360 |
0.25 |
| chr11_60199988_60200509 | 1.12 |
Mir6921 |
microRNA 6921 |
369 |
0.65 |
| chr3_106482771_106483037 | 1.12 |
Dennd2d |
DENN/MADD domain containing 2D |
454 |
0.49 |
| chr11_46085325_46085478 | 1.11 |
Mir8100 |
microRNA 8100 |
16869 |
0.12 |
| chr10_127373842_127374054 | 1.10 |
Inhbc |
inhibin beta-C |
3517 |
0.11 |
| chr2_150786904_150787055 | 1.09 |
Pygb |
brain glycogen phosphorylase |
244 |
0.91 |
| chr8_70551140_70551291 | 1.08 |
Ell |
elongation factor RNA polymerase II |
11527 |
0.08 |
| chr17_24163699_24163861 | 1.05 |
Amdhd2 |
amidohydrolase domain containing 2 |
14 |
0.94 |
| chr9_106238027_106238241 | 1.05 |
Alas1 |
aminolevulinic acid synthase 1 |
596 |
0.58 |
| chr5_147071140_147071411 | 1.03 |
Lnx2 |
ligand of numb-protein X 2 |
5311 |
0.17 |
| chr2_84840241_84840491 | 1.03 |
Slc43a1 |
solute carrier family 43, member 1 |
167 |
0.91 |
| chr12_29894783_29894948 | 1.03 |
Myt1l |
myelin transcription factor 1-like |
249 |
0.95 |
| chr12_103328324_103328502 | 1.02 |
Asb2 |
ankyrin repeat and SOCS box-containing 2 |
7171 |
0.11 |
| chr9_94705088_94705258 | 1.01 |
Gm16262 |
predicted gene 16262 |
2854 |
0.25 |
| chr19_20610921_20611108 | 1.00 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
9053 |
0.22 |
| chr10_128909533_128909836 | 1.00 |
Cd63 |
CD63 antigen |
15 |
0.95 |
| chr11_5899422_5899614 | 0.99 |
Myl7 |
myosin, light polypeptide 7, regulatory |
736 |
0.5 |
| chr17_87570754_87570962 | 0.98 |
Gm46587 |
predicted gene, 46587 |
796 |
0.62 |
| chr11_98754743_98754913 | 0.98 |
Thra |
thyroid hormone receptor alpha |
1242 |
0.28 |
| chr2_168721341_168721659 | 0.97 |
Atp9a |
ATPase, class II, type 9A |
8538 |
0.2 |
| chr2_155067847_155068055 | 0.97 |
Gm45609 |
predicted gene 45609 |
6230 |
0.14 |
| chr10_59149959_59150116 | 0.96 |
Septin10 |
septin 10 |
28820 |
0.16 |
| chr9_78154656_78154974 | 0.95 |
Cilk1 |
ciliogenesis associated kinase 1 |
508 |
0.69 |
| chr6_139577165_139577358 | 0.94 |
4930443G03Rik |
RIKEN cDNA 4930443G03 gene |
56 |
0.98 |
| chr3_94412685_94412888 | 0.94 |
1700040D17Rik |
RIKEN cDNA 1700040D17 gene |
133 |
0.72 |
| chr4_137795816_137796289 | 0.94 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
178 |
0.96 |
| chr4_149978768_149979375 | 0.93 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
22194 |
0.13 |
| chr17_28517484_28517972 | 0.93 |
Fkbp5 |
FK506 binding protein 5 |
201 |
0.84 |
| chr17_24960376_24960530 | 0.89 |
Jpt2 |
Jupiter microtubule associated homolog 2 |
183 |
0.91 |
| chr9_24420554_24420746 | 0.88 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
3309 |
0.28 |
| chr5_139427163_139427327 | 0.88 |
Gper1 |
G protein-coupled estrogen receptor 1 |
3965 |
0.14 |
| chr1_190822997_190823215 | 0.87 |
Rps6kc1 |
ribosomal protein S6 kinase polypeptide 1 |
10930 |
0.24 |
| chr4_148168418_148168586 | 0.86 |
Fbxo2 |
F-box protein 2 |
7881 |
0.11 |
| chr5_137641169_137641333 | 0.85 |
Sap25 |
sin3 associated polypeptide |
83 |
0.9 |
| chr18_76242274_76242708 | 0.85 |
Smad2 |
SMAD family member 2 |
317 |
0.89 |
| chr11_117780625_117780787 | 0.85 |
Tmc6 |
transmembrane channel-like gene family 6 |
48 |
0.93 |
| chr7_101085910_101086353 | 0.84 |
Fchsd2 |
FCH and double SH3 domains 2 |
6732 |
0.17 |
| chr2_72157539_72157839 | 0.84 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
8893 |
0.2 |
| chr10_62833072_62833249 | 0.84 |
Tet1 |
tet methylcytosine dioxygenase 1 |
18619 |
0.11 |
| chr12_28700387_28700829 | 0.84 |
Trappc12 |
trafficking protein particle complex 12 |
1001 |
0.46 |
| chr11_5909915_5910066 | 0.83 |
Gck |
glucokinase |
5134 |
0.12 |
| chr14_118040435_118040621 | 0.83 |
Dct |
dopachrome tautomerase |
2601 |
0.27 |
| chr10_63339095_63339253 | 0.83 |
Sirt1 |
sirtuin 1 |
139 |
0.94 |
| chr19_23074772_23074923 | 0.83 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
1006 |
0.54 |
| chr1_75142621_75142779 | 0.83 |
Cnppd1 |
cyclin Pas1/PHO80 domain containing 1 |
3 |
0.54 |
| chr8_80821631_80821838 | 0.82 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
9150 |
0.2 |
| chr8_105798853_105799004 | 0.82 |
Ranbp10 |
RAN binding protein 10 |
28277 |
0.08 |
| chr3_132815131_132815356 | 0.82 |
Gm42876 |
predicted gene 42876 |
8990 |
0.14 |
| chr5_4960222_4960441 | 0.82 |
Gm22897 |
predicted gene, 22897 |
24906 |
0.18 |
| chr11_117341816_117342087 | 0.79 |
Septin9 |
septin 9 |
9523 |
0.19 |
| chr7_141293897_141294068 | 0.79 |
Drd4 |
dopamine receptor D4 |
851 |
0.35 |
| chr19_20726917_20727068 | 0.78 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
570 |
0.83 |
| chr6_108140829_108140991 | 0.78 |
Rpl36-ps12 |
ribosomal protein L36, pseudogene 12 |
401 |
0.86 |
| chr10_80340392_80340718 | 0.77 |
Adamtsl5 |
ADAMTS-like 5 |
470 |
0.56 |
| chr9_74850759_74851075 | 0.77 |
Gm16551 |
predicted gene 16551 |
1922 |
0.24 |
| chr6_140368798_140368983 | 0.77 |
n-R5s168 |
nuclear encoded rRNA 5S 168 |
49260 |
0.11 |
| chr16_97913947_97914098 | 0.77 |
C2cd2 |
C2 calcium-dependent domain containing 2 |
8584 |
0.18 |
| chr5_66135628_66135793 | 0.76 |
Rbm47 |
RNA binding motif protein 47 |
15246 |
0.11 |
| chr7_24928137_24928288 | 0.76 |
Arhgef1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
4849 |
0.1 |
| chr1_105998118_105998303 | 0.75 |
Zcchc2 |
zinc finger, CCHC domain containing 2 |
5912 |
0.17 |
| chr1_51878776_51878948 | 0.74 |
Gm28323 |
predicted gene 28323 |
746 |
0.61 |
| chr9_113122162_113122366 | 0.74 |
Gm36251 |
predicted gene, 36251 |
765 |
0.76 |
| chr11_28698214_28698534 | 0.74 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
16810 |
0.16 |
| chr4_148038831_148039191 | 0.74 |
Mthfr |
methylenetetrahydrofolate reductase |
66 |
0.66 |
| chr18_75675824_75676322 | 0.73 |
Ctif |
CBP80/20-dependent translation initiation factor |
21481 |
0.23 |
| chr4_144917193_144917364 | 0.73 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
1601 |
0.41 |
| chr17_28812253_28812687 | 0.73 |
Brpf3 |
bromodomain and PHD finger containing, 3 |
1919 |
0.2 |
| chr1_171195959_171196110 | 0.73 |
Pcp4l1 |
Purkinje cell protein 4-like 1 |
234 |
0.84 |
| chr2_27572911_27573062 | 0.73 |
Gm13421 |
predicted gene 13421 |
32557 |
0.13 |
| chr7_123196570_123196734 | 0.72 |
Tnrc6a |
trinucleotide repeat containing 6a |
16731 |
0.19 |
| chr11_70000432_70000600 | 0.71 |
Dvl2 |
dishevelled segment polarity protein 2 |
79 |
0.9 |
| chr19_44405361_44405580 | 0.71 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
1220 |
0.39 |
| chr10_78426542_78426693 | 0.70 |
Cstb |
cystatin B |
948 |
0.28 |
| chr6_90568565_90568716 | 0.70 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
934 |
0.49 |
| chr10_117145636_117145926 | 0.70 |
Frs2 |
fibroblast growth factor receptor substrate 2 |
2714 |
0.21 |
| chr2_173159829_173160580 | 0.69 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
7122 |
0.18 |
| chr8_120113112_120113406 | 0.69 |
6430548M08Rik |
RIKEN cDNA 6430548M08 gene |
893 |
0.44 |
| chr19_44985600_44985928 | 0.69 |
4930414N06Rik |
RIKEN cDNA 4930414N06 gene |
1214 |
0.29 |
| chr11_60196530_60196694 | 0.69 |
Mir6921 |
microRNA 6921 |
4005 |
0.13 |
| chr8_111854325_111854488 | 0.69 |
Cfdp1 |
craniofacial development protein 1 |
115 |
0.96 |
| chr9_102732818_102732969 | 0.69 |
Amotl2 |
angiomotin-like 2 |
2162 |
0.21 |
| chr11_75769198_75769770 | 0.68 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
17880 |
0.14 |
| chr15_83349552_83350074 | 0.68 |
Arfgap3 |
ADP-ribosylation factor GTPase activating protein 3 |
361 |
0.84 |
| chr8_41069792_41069999 | 0.68 |
Mtus1 |
mitochondrial tumor suppressor 1 |
12881 |
0.16 |
| chr12_103949173_103949487 | 0.68 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
7568 |
0.11 |
| chr3_157912600_157912930 | 0.67 |
Cth |
cystathionase (cystathionine gamma-lyase) |
7040 |
0.14 |
| chr10_20148225_20148689 | 0.67 |
Map7 |
microtubule-associated protein 7 |
14 |
0.98 |
| chr14_59200883_59201288 | 0.67 |
Rcbtb1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
124 |
0.96 |
| chr8_116268180_116268552 | 0.67 |
4930422C21Rik |
RIKEN cDNA 4930422C21 gene |
80997 |
0.1 |
| chr11_51584956_51585449 | 0.67 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
391 |
0.8 |
| chr3_152396303_152396479 | 0.66 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
259 |
0.84 |
| chr17_31521089_31521268 | 0.66 |
Ndufv3 |
NADH:ubiquinone oxidoreductase core subunit V3 |
993 |
0.26 |
| chr9_58582090_58582367 | 0.66 |
Nptn |
neuroplastin |
12 |
0.98 |
| chr12_55831200_55831355 | 0.66 |
Brms1l |
breast cancer metastasis-suppressor 1-like |
5047 |
0.15 |
| chr7_19877096_19877247 | 0.66 |
Gm44659 |
predicted gene 44659 |
5733 |
0.08 |
| chr3_133544844_133545488 | 0.65 |
Tet2 |
tet methylcytosine dioxygenase 2 |
27 |
0.53 |
| chr15_76227819_76227970 | 0.65 |
Plec |
plectin |
1603 |
0.17 |
| chr11_81370078_81370386 | 0.64 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
14781 |
0.29 |
| chr5_121338798_121338949 | 0.64 |
Gm24671 |
predicted gene, 24671 |
17971 |
0.11 |
| chr12_84970404_84970739 | 0.64 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
253 |
0.56 |
| chr6_128438925_128439109 | 0.64 |
Gm10069 |
predicted gene 10069 |
260 |
0.53 |
| chr2_144369139_144369688 | 0.64 |
Kat14 |
lysine acetyltransferase 14 |
414 |
0.43 |
| chr5_9032304_9032484 | 0.63 |
Gm40264 |
predicted gene, 40264 |
2386 |
0.22 |
| chr11_120627879_120628086 | 0.62 |
Mafg |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
2166 |
0.09 |
| chr11_119258845_119259008 | 0.62 |
Ccdc40 |
coiled-coil domain containing 40 |
1 |
0.96 |
| chr14_70443158_70443325 | 0.62 |
Polr3d |
polymerase (RNA) III (DNA directed) polypeptide D |
49 |
0.76 |
| chr17_56233947_56234106 | 0.61 |
Gm49879 |
predicted gene, 49879 |
4883 |
0.1 |
| chr9_57439182_57439333 | 0.61 |
Ppcdc |
phosphopantothenoylcysteine decarboxylase |
834 |
0.49 |
| chr17_86144597_86145055 | 0.61 |
Srbd1 |
S1 RNA binding domain 1 |
349 |
0.89 |
| chr16_17922186_17922530 | 0.61 |
AA914427 |
EST AA914427 |
315 |
0.77 |
| chr18_60243095_60243283 | 0.60 |
Gm5970 |
predicted gene 5970 |
22346 |
0.13 |
| chr13_90089026_90089404 | 0.60 |
Tmem167 |
transmembrane protein 167 |
92 |
0.56 |
| chr8_123232613_123232964 | 0.60 |
Gm45842 |
predicted gene 45842 |
1989 |
0.12 |
| chr13_43252482_43252652 | 0.59 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
50838 |
0.13 |
| chr15_34238004_34238174 | 0.59 |
Laptm4b |
lysosomal-associated protein transmembrane 4B |
60 |
0.97 |
| chr10_68094854_68095005 | 0.58 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
41697 |
0.15 |
| chr9_87255292_87255453 | 0.58 |
Cep162 |
centrosomal protein 162 |
84 |
0.98 |
| chr2_31477009_31477347 | 0.58 |
Ass1 |
argininosuccinate synthetase 1 |
6971 |
0.19 |
| chr2_164073997_164074164 | 0.58 |
Stk4 |
serine/threonine kinase 4 |
58 |
0.96 |
| chr5_137314174_137314330 | 0.58 |
Trip6 |
thyroid hormone receptor interactor 6 |
45 |
0.93 |
| chr13_51946235_51946429 | 0.58 |
Gm26651 |
predicted gene, 26651 |
26882 |
0.2 |
| chr9_120492375_120492774 | 0.57 |
D830035M03Rik |
RIKEN cDNA D830035M03 gene |
27 |
0.55 |
| chr14_70351384_70351576 | 0.57 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
55 |
0.96 |
| chr11_62557761_62557924 | 0.57 |
Ubb |
ubiquitin B |
6166 |
0.09 |
| chr3_21936262_21936525 | 0.55 |
Gm43674 |
predicted gene 43674 |
62075 |
0.12 |
| chr9_103761868_103762039 | 0.55 |
Tmem108 |
transmembrane protein 108 |
116 |
0.98 |
| chr10_99209224_99209646 | 0.55 |
Gm34574 |
predicted gene, 34574 |
13631 |
0.11 |
| chr6_9078648_9078799 | 0.55 |
Gm35736 |
predicted gene, 35736 |
68043 |
0.14 |
| chr2_121449025_121449188 | 0.54 |
Serf2 |
small EDRK-rich factor 2 |
89 |
0.9 |
| chr6_8209868_8210031 | 0.54 |
Mios |
meiosis regulator for oocyte development |
727 |
0.65 |
| chr4_154864284_154864460 | 0.54 |
Ttc34 |
tetratricopeptide repeat domain 34 |
3820 |
0.16 |
| chr2_167496630_167496810 | 0.53 |
Spata2 |
spermatogenesis associated 2 |
3833 |
0.16 |
| chr2_180725008_180725370 | 0.53 |
Slc17a9 |
solute carrier family 17, member 9 |
74 |
0.95 |
| chr13_21171668_21171974 | 0.53 |
Trim27 |
tripartite motif-containing 27 |
7624 |
0.14 |
| chr17_22361357_22361742 | 0.53 |
Zfp758 |
zinc finger protein 758 |
27 |
0.65 |
| chr3_40540672_40540830 | 0.53 |
Intu |
inturned planar cell polarity protein |
16 |
0.98 |
| chr11_69088418_69088577 | 0.53 |
Vamp2 |
vesicle-associated membrane protein 2 |
7 |
0.93 |
| chr19_25542114_25542287 | 0.52 |
Dmrt1 |
doublesex and mab-3 related transcription factor 1 |
36493 |
0.15 |
| chr10_95685288_95685475 | 0.52 |
Anapc15-ps |
anaphase promoting complex C subunit 15, pseudogene |
11930 |
0.11 |
| chr5_110102038_110102206 | 0.52 |
Plcxd1 |
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
918 |
0.36 |
| chr4_109419168_109419344 | 0.51 |
Ttc39a |
tetratricopeptide repeat domain 39A |
1686 |
0.32 |
| chr3_116236671_116236856 | 0.51 |
Gpr88 |
G-protein coupled receptor 88 |
16326 |
0.17 |
| chr5_72168137_72168298 | 0.51 |
Commd8 |
COMM domain containing 8 |
28 |
0.98 |
| chr9_63989489_63989681 | 0.51 |
Smad6 |
SMAD family member 6 |
27402 |
0.13 |
| chr17_80625437_80625624 | 0.50 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
544 |
0.81 |
| chr2_170007586_170007998 | 0.50 |
AY702102 |
cDNA sequence AY702102 |
44605 |
0.18 |
| chr7_123186591_123187282 | 0.50 |
Tnrc6a |
trinucleotide repeat containing 6a |
7015 |
0.22 |
| chr16_84955565_84955728 | 0.50 |
App |
amyloid beta (A4) precursor protein |
7735 |
0.18 |
| chr7_128617191_128617383 | 0.50 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
5927 |
0.13 |
| chr3_138221221_138221499 | 0.50 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
111 |
0.95 |
| chr5_123172881_123173032 | 0.50 |
Hpd |
4-hydroxyphenylpyruvic acid dioxygenase |
45 |
0.94 |
| chrX_42013544_42013707 | 0.50 |
Xiap |
X-linked inhibitor of apoptosis |
46054 |
0.13 |
| chr14_58788638_58788797 | 0.49 |
Gm9022 |
predicted gene 9022 |
90598 |
0.08 |
| chrX_163908846_163909025 | 0.49 |
Ap1s2 |
adaptor-related protein complex 1, sigma 2 subunit |
82 |
0.98 |
| chr11_94653911_94654072 | 0.49 |
Mrpl27 |
mitochondrial ribosomal protein L27 |
20 |
0.49 |
| chr6_52172625_52172818 | 0.49 |
Gm15050 |
predicted gene 15050 |
160 |
0.83 |
| chr9_106369294_106369462 | 0.49 |
Dusp7 |
dual specificity phosphatase 7 |
234 |
0.89 |
| chr15_85337324_85337483 | 0.48 |
Atxn10 |
ataxin 10 |
1158 |
0.51 |
| chr9_53596608_53596772 | 0.48 |
Acat1 |
acetyl-Coenzyme A acetyltransferase 1 |
12933 |
0.14 |
| chr1_134419856_134420043 | 0.48 |
Adipor1 |
adiponectin receptor 1 |
4385 |
0.12 |
| chr11_117072260_117072440 | 0.48 |
Snhg20 |
small nucleolar RNA host gene 20 |
3658 |
0.12 |
| chr5_74401642_74401799 | 0.48 |
Scfd2 |
Sec1 family domain containing 2 |
3982 |
0.2 |
| chr13_23526499_23526651 | 0.48 |
Gm11335 |
predicted gene 11335 |
4368 |
0.06 |
| chr13_47014530_47014726 | 0.48 |
Nhlrc1 |
NHL repeat containing 1 |
222 |
0.86 |
| chr2_17644701_17644889 | 0.48 |
Mir6419 |
microRNA 6419 |
81281 |
0.09 |
| chr13_101746510_101746927 | 0.48 |
Gm36638 |
predicted gene, 36638 |
5170 |
0.24 |
| chr6_115804879_115805037 | 0.48 |
Snora7a |
small nucleolar RNA, H/ACA box 7A |
3156 |
0.12 |
| chr18_5010031_5010219 | 0.47 |
Svil |
supervillin |
15189 |
0.28 |
| chr7_111122949_111123134 | 0.47 |
1700012D14Rik |
RIKEN cDNA 1700012D14 gene |
357 |
0.84 |
| chr6_101377687_101377878 | 0.47 |
Pdzrn3 |
PDZ domain containing RING finger 3 |
115 |
0.96 |
| chr13_23504004_23504163 | 0.47 |
n-TStga1 |
nuclear encoded tRNA serine 1 (anticodon TGA) |
14059 |
0.05 |
| chr6_118479189_118479349 | 0.46 |
Zfp9 |
zinc finger protein 9 |
51 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.3 | 0.8 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.3 | 0.8 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.3 | 1.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.2 | 0.9 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 0.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 1.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.3 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.2 | 0.5 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.2 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.8 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 1.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.4 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.5 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 0.4 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.4 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.5 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.3 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.1 | 0.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.1 | 0.2 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 1.0 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.4 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.4 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.2 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.2 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.4 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0006548 | histidine metabolic process(GO:0006547) histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.3 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 1.0 | GO:0051304 | chromosome separation(GO:0051304) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.1 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.0 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0040033 | negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.0 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.0 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.4 | GO:1904029 | regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.3 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 0.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 0.8 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0080031 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0044466 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.1 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0070635 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.6 | GO:0042281 | dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042281) |
| 0.0 | 2.0 | GO:0008766 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME CELL CYCLE CHECKPOINTS | Genes involved in Cell Cycle Checkpoints |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |