Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
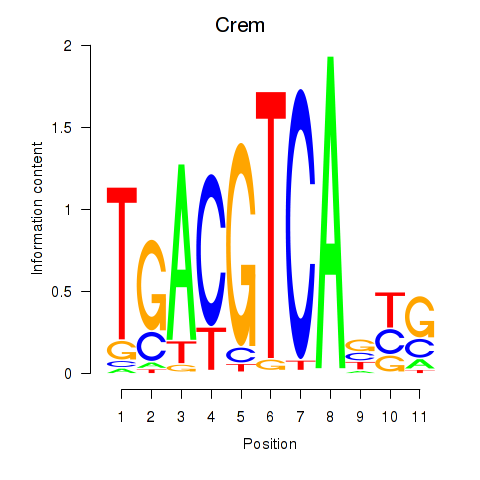
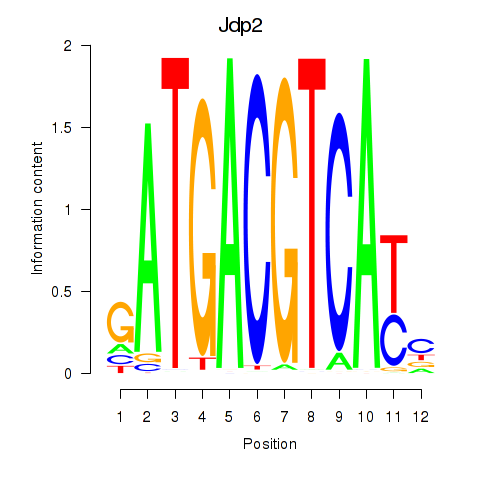
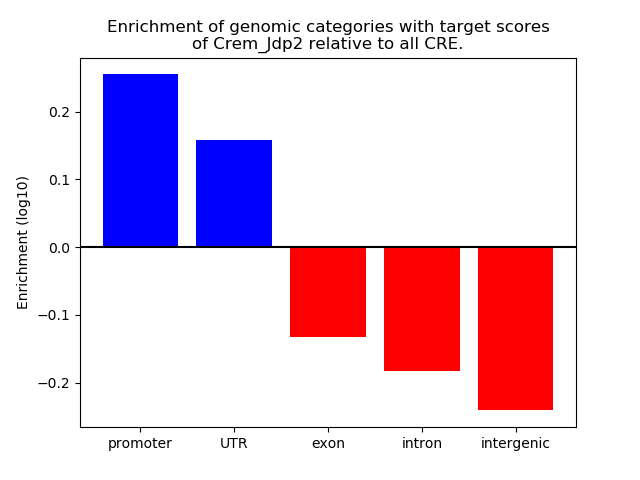
Results for Crem_Jdp2
Z-value: 2.47
Transcription factors associated with Crem_Jdp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Crem
|
ENSMUSG00000063889.10 | cAMP responsive element modulator |
|
Jdp2
|
ENSMUSG00000034271.9 | Jun dimerization protein 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_3269297_3269536 | Crem | 11662 | 0.216117 | 0.80 | 5.6e-02 | Click! |
| chr18_3269107_3269266 | Crem | 11892 | 0.215604 | 0.74 | 9.5e-02 | Click! |
| chr18_3279745_3279919 | Crem | 1246 | 0.514834 | 0.73 | 9.8e-02 | Click! |
| chr18_3337336_3337491 | Crem | 75 | 0.919774 | -0.67 | 1.4e-01 | Click! |
| chr18_3281039_3281202 | Crem | 12 | 0.982054 | -0.64 | 1.7e-01 | Click! |
| chr12_85598845_85598996 | Jdp2 | 107 | 0.718457 | 0.79 | 6.4e-02 | Click! |
| chr12_85599222_85599387 | Jdp2 | 112 | 0.493118 | -0.69 | 1.3e-01 | Click! |
| chr12_85598582_85598751 | Jdp2 | 361 | 0.597353 | 0.67 | 1.5e-01 | Click! |
| chr12_85598165_85598354 | Jdp2 | 768 | 0.443604 | -0.58 | 2.2e-01 | Click! |
| chr12_85620383_85620537 | Jdp2 | 18238 | 0.138236 | 0.43 | 4.0e-01 | Click! |
Activity of the Crem_Jdp2 motif across conditions
Conditions sorted by the z-value of the Crem_Jdp2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
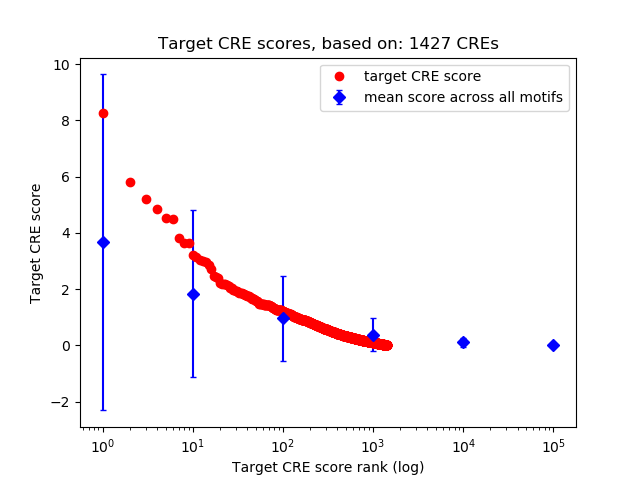
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_61226929_61227456 | 8.27 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr3_51255928_51256079 | 5.80 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr11_75173089_75173918 | 5.21 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr7_97788226_97788577 | 4.84 |
Pak1 |
p21 (RAC1) activated kinase 1 |
140 |
0.96 |
| chr7_112679198_112679362 | 4.53 |
Tead1 |
TEA domain family member 1 |
38 |
0.94 |
| chr10_17948008_17948419 | 4.51 |
Heca |
hdc homolog, cell cycle regulator |
146 |
0.97 |
| chr14_61680870_61681697 | 3.83 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr7_142095283_142095608 | 3.64 |
Dusp8 |
dual specificity phosphatase 8 |
173 |
0.64 |
| chr19_61226381_61226532 | 3.63 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
227 |
0.89 |
| chr14_66103708_66103859 | 3.22 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
6877 |
0.17 |
| chr14_120477462_120478025 | 3.14 |
Rap2a |
RAS related protein 2a |
701 |
0.77 |
| chr2_79448009_79448160 | 3.03 |
Neurod1 |
neurogenic differentiation 1 |
8667 |
0.21 |
| chr16_78306591_78306742 | 3.02 |
E330011O21Rik |
RIKEN cDNA E330011O21 gene |
4179 |
0.18 |
| chr2_72741614_72741840 | 2.95 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
9126 |
0.22 |
| chr5_124032266_124032428 | 2.87 |
Gm34086 |
predicted gene, 34086 |
14 |
0.54 |
| chr12_59219708_59220018 | 2.72 |
Fbxo33 |
F-box protein 33 |
138 |
0.95 |
| chr17_27820677_27821072 | 2.45 |
Ilrun |
inflammation and lipid regulator with UBA-like and NBR1-like domains |
226 |
0.89 |
| chr13_53001712_53001870 | 2.44 |
Gm33424 |
predicted gene, 33424 |
14411 |
0.16 |
| chr11_103102027_103102382 | 2.38 |
Acbd4 |
acyl-Coenzyme A binding domain containing 4 |
478 |
0.46 |
| chr7_49459741_49459913 | 2.21 |
Nav2 |
neuron navigator 2 |
9067 |
0.23 |
| chr4_134245531_134245693 | 2.18 |
Zfp593 |
zinc finger protein 593 |
20 |
0.92 |
| chr2_180589607_180589819 | 2.17 |
Ogfr |
opioid growth factor receptor |
251 |
0.89 |
| chr8_105606555_105606946 | 2.17 |
Ripor1 |
RHO family interacting cell polarization regulator 1 |
1495 |
0.2 |
| chr6_39725511_39725691 | 2.15 |
Braf |
Braf transforming gene |
138 |
0.96 |
| chr2_11706768_11706947 | 2.10 |
Il15ra |
interleukin 15 receptor, alpha chain |
1009 |
0.47 |
| chr8_122775635_122775811 | 2.04 |
Acsf3 |
acyl-CoA synthetase family member 3 |
145 |
0.68 |
| chr10_82240847_82241161 | 2.03 |
Zfp938 |
zinc finger protein 938 |
269 |
0.89 |
| chr15_58941890_58942087 | 1.98 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
8180 |
0.13 |
| chr15_12321006_12321530 | 1.96 |
Golph3 |
golgi phosphoprotein 3 |
182 |
0.84 |
| chr14_76827348_76827510 | 1.95 |
Gm48968 |
predicted gene, 48968 |
5392 |
0.22 |
| chr13_97775135_97775301 | 1.92 |
Rps18-ps6 |
ribosomal protein S18, pseudogene 6 |
14586 |
0.14 |
| chr8_111630041_111630248 | 1.91 |
Ldhd |
lactate dehydrogenase D |
191 |
0.93 |
| chr12_111758755_111758932 | 1.87 |
Klc1 |
kinesin light chain 1 |
6 |
0.96 |
| chr6_14900560_14901049 | 1.85 |
Foxp2 |
forkhead box P2 |
545 |
0.87 |
| chr10_87926267_87926684 | 1.84 |
Tyms-ps |
thymidylate synthase, pseudogene |
40372 |
0.13 |
| chr18_73844340_73844698 | 1.84 |
Mro |
maestro |
14866 |
0.2 |
| chr12_84217297_84217462 | 1.82 |
Gm47447 |
predicted gene, 47447 |
1242 |
0.27 |
| chr19_43463068_43463222 | 1.81 |
Gm47938 |
predicted gene, 47938 |
6613 |
0.14 |
| chr10_80622017_80622180 | 1.78 |
Csnk1g2 |
casein kinase 1, gamma 2 |
740 |
0.4 |
| chr4_88868228_88868464 | 1.77 |
4930553M12Rik |
RIKEN cDNA 4930553M12 gene |
33 |
0.93 |
| chr18_74939611_74939786 | 1.76 |
Gm18786 |
predicted gene, 18786 |
3418 |
0.13 |
| chr8_70500942_70501204 | 1.76 |
Crlf1 |
cytokine receptor-like factor 1 |
60 |
0.93 |
| chr14_67072405_67072747 | 1.70 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
132 |
0.95 |
| chr2_58756221_58756376 | 1.68 |
Upp2 |
uridine phosphorylase 2 |
1086 |
0.54 |
| chr10_95616497_95616648 | 1.67 |
Gm33336 |
predicted gene, 33336 |
15667 |
0.12 |
| chr1_53313553_53313713 | 1.66 |
Osgepl1 |
O-sialoglycoprotein endopeptidase-like 1 |
9 |
0.95 |
| chr13_95639510_95639665 | 1.64 |
F2r |
coagulation factor II (thrombin) receptor |
21100 |
0.13 |
| chr17_47441317_47441743 | 1.64 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
4154 |
0.13 |
| chr7_98389139_98389290 | 1.62 |
Gm44507 |
predicted gene 44507 |
26410 |
0.12 |
| chr4_137919760_137920083 | 1.58 |
Ece1 |
endothelin converting enzyme 1 |
6239 |
0.22 |
| chr15_7175278_7175429 | 1.58 |
Lifr |
LIF receptor alpha |
21000 |
0.23 |
| chr19_34524558_34524859 | 1.58 |
Lipa |
lysosomal acid lipase A |
2703 |
0.19 |
| chr15_79347426_79347596 | 1.56 |
Maff |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
9 |
0.96 |
| chr5_150522352_150522676 | 1.49 |
Brca2 |
breast cancer 2, early onset |
116 |
0.92 |
| chr3_138288275_138288480 | 1.49 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
10726 |
0.12 |
| chr3_153250704_153250860 | 1.49 |
St6galnac3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
24999 |
0.17 |
| chr3_145118680_145119047 | 1.48 |
Odf2l |
outer dense fiber of sperm tails 2-like |
111 |
0.97 |
| chr17_26508364_26508746 | 1.47 |
Dusp1 |
dual specificity phosphatase 1 |
36 |
0.96 |
| chr11_16850729_16850984 | 1.46 |
Egfros |
epidermal growth factor receptor, opposite strand |
20154 |
0.18 |
| chr19_17617854_17618016 | 1.45 |
Gm17819 |
predicted gene, 17819 |
74828 |
0.11 |
| chr5_110652988_110653519 | 1.45 |
Noc4l |
NOC4 like |
6 |
0.71 |
| chr11_16852969_16853170 | 1.45 |
Egfros |
epidermal growth factor receptor, opposite strand |
22367 |
0.17 |
| chr6_17584009_17584205 | 1.44 |
Met |
met proto-oncogene |
37134 |
0.15 |
| chr4_149398701_149398906 | 1.44 |
Ube4b |
ubiquitination factor E4B |
575 |
0.68 |
| chr1_75479439_75479625 | 1.44 |
Tmem198 |
transmembrane protein 198 |
0 |
0.73 |
| chr15_38373856_38374248 | 1.44 |
Gm41307 |
predicted gene, 41307 |
30102 |
0.13 |
| chr13_114083297_114083584 | 1.43 |
Gm47479 |
predicted gene, 47479 |
36976 |
0.18 |
| chr15_79455529_79455727 | 1.43 |
Csnk1e |
casein kinase 1, epsilon |
62 |
0.95 |
| chr3_146630472_146630703 | 1.42 |
Gm16325 |
predicted gene 16325 |
11345 |
0.12 |
| chr17_53478848_53479280 | 1.42 |
Rab5a |
RAB5A, member RAS oncogene family |
170 |
0.93 |
| chr16_4077809_4077973 | 1.42 |
Trap1 |
TNF receptor-associated protein 1 |
64 |
0.95 |
| chr18_60243665_60243848 | 1.41 |
Gm5970 |
predicted gene 5970 |
22913 |
0.13 |
| chr5_105198920_105199230 | 1.41 |
Gbp10 |
guanylate-binding protein 10 |
22901 |
0.16 |
| chr3_89091548_89091781 | 1.41 |
Rusc1 |
RUN and SH3 domain containing 1 |
1619 |
0.18 |
| chr13_17837836_17838002 | 1.35 |
Gm18859 |
predicted gene, 18859 |
16973 |
0.12 |
| chr4_45505286_45505685 | 1.35 |
Gm22518 |
predicted gene, 22518 |
4933 |
0.17 |
| chr2_156065132_156065865 | 1.34 |
Spag4 |
sperm associated antigen 4 |
55 |
0.94 |
| chr2_155070811_155070993 | 1.34 |
Gm45609 |
predicted gene 45609 |
3279 |
0.17 |
| chr2_15055062_15055234 | 1.32 |
Nsun6 |
NOL1/NOP2/Sun domain family member 6 |
79 |
0.51 |
| chrY_90783982_90784262 | 1.31 |
Gm47283 |
predicted gene, 47283 |
616 |
0.72 |
| chr18_66458471_66458640 | 1.30 |
Pmaip1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
20 |
0.96 |
| chr1_59237644_59237808 | 1.29 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
495 |
0.78 |
| chr3_32418021_32418172 | 1.28 |
Pik3ca |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
18055 |
0.15 |
| chr9_69397707_69397993 | 1.27 |
Ice2 |
interactor of little elongation complex ELL subunit 2 |
56 |
0.97 |
| chr16_5049903_5050066 | 1.27 |
Ubn1 |
ubinuclein 1 |
73 |
0.56 |
| chr15_101453443_101453620 | 1.27 |
Krt88 |
keratin 88 |
5787 |
0.08 |
| chr1_79439965_79440133 | 1.27 |
Scg2 |
secretogranin II |
7 |
0.98 |
| chr13_67360549_67360708 | 1.27 |
Zfp953 |
zinc finger protein 953 |
23 |
0.36 |
| chr9_55264900_55265092 | 1.26 |
Nrg4 |
neuregulin 4 |
18576 |
0.16 |
| chr18_74958532_74958840 | 1.26 |
Lipg |
lipase, endothelial |
2576 |
0.14 |
| chr10_76997859_76998018 | 1.26 |
Gm35721 |
predicted gene, 35721 |
10631 |
0.12 |
| chr9_43224415_43225053 | 1.26 |
Oaf |
out at first homolog |
358 |
0.84 |
| chr17_24168769_24169218 | 1.25 |
Atp6v0c |
ATPase, H+ transporting, lysosomal V0 subunit C |
73 |
0.93 |
| chr7_118584670_118584840 | 1.25 |
Tmc7 |
transmembrane channel-like gene family 7 |
19 |
0.97 |
| chr17_46890284_46890668 | 1.25 |
Tbcc |
tubulin-specific chaperone C |
208 |
0.92 |
| chr3_96115042_96115201 | 1.25 |
Otud7b |
OTU domain containing 7B |
8034 |
0.09 |
| chr17_27907441_27908078 | 1.23 |
Taf11 |
TATA-box binding protein associated factor 11 |
24 |
0.5 |
| chr1_24678416_24678721 | 1.21 |
Lmbrd1 |
LMBR1 domain containing 1 |
62 |
0.97 |
| chr6_144256546_144256717 | 1.21 |
Sox5 |
SRY (sex determining region Y)-box 5 |
47063 |
0.19 |
| chr5_116425330_116425492 | 1.21 |
Hspb8 |
heat shock protein 8 |
2547 |
0.19 |
| chr5_134676027_134676369 | 1.21 |
Gm10369 |
predicted gene 10369 |
292 |
0.84 |
| chr13_100104015_100104175 | 1.20 |
Bdp1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
25 |
0.97 |
| chr3_130635863_130636014 | 1.19 |
Etnppl |
ethanolamine phosphate phospholyase |
9418 |
0.16 |
| chr2_31476693_31476999 | 1.19 |
Ass1 |
argininosuccinate synthetase 1 |
6639 |
0.2 |
| chr12_86884532_86884993 | 1.18 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
36 |
0.98 |
| chr8_111145184_111145817 | 1.18 |
9430091E24Rik |
RIKEN cDNA 9430091E24 gene |
20 |
0.97 |
| chr13_67813932_67814279 | 1.17 |
Zfp273 |
zinc finger protein 273 |
289 |
0.81 |
| chr2_155276592_155276746 | 1.16 |
Map1lc3a |
microtubule-associated protein 1 light chain 3 alpha |
126 |
0.96 |
| chr1_166409779_166409955 | 1.16 |
Pogk |
pogo transposable element with KRAB domain |
4 |
0.97 |
| chr6_30304499_30304656 | 1.15 |
1700095J07Rik |
RIKEN cDNA 1700095J07 gene |
28 |
0.5 |
| chr15_33687773_33687947 | 1.15 |
Tspyl5 |
testis-specific protein, Y-encoded-like 5 |
24 |
0.98 |
| chr6_84698036_84698213 | 1.14 |
Cyp26b1 |
cytochrome P450, family 26, subfamily b, polypeptide 1 |
104216 |
0.06 |
| chr2_52603393_52603554 | 1.13 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
16281 |
0.2 |
| chr6_90599935_90600097 | 1.13 |
Gm15756 |
predicted gene 15756 |
68 |
0.96 |
| chr15_62037707_62038323 | 1.12 |
Pvt1 |
Pvt1 oncogene |
23 |
0.98 |
| chr3_67373939_67374117 | 1.12 |
Mlf1 |
myeloid leukemia factor 1 |
69 |
0.95 |
| chr5_122059761_122059912 | 1.11 |
Cux2 |
cut-like homeobox 2 |
9734 |
0.14 |
| chrX_170009083_170009242 | 1.11 |
Erdr1 |
erythroid differentiation regulator 1 |
497 |
0.77 |
| chr3_102677050_102677222 | 1.11 |
Gm19202 |
predicted gene, 19202 |
9581 |
0.14 |
| chr2_122380837_122380998 | 1.11 |
Gm24409 |
predicted gene, 24409 |
4723 |
0.15 |
| chr19_55268150_55268498 | 1.10 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
4546 |
0.2 |
| chr9_96478340_96478816 | 1.10 |
Rnf7 |
ring finger protein 7 |
10 |
0.97 |
| chr5_120139175_120139351 | 1.09 |
Gm10390 |
predicted gene 10390 |
0 |
0.98 |
| chr2_24179409_24179581 | 1.09 |
Gm13410 |
predicted gene 13410 |
661 |
0.62 |
| chr13_95858927_95859107 | 1.08 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
32740 |
0.16 |
| chr8_122306649_122307232 | 1.08 |
Zfpm1 |
zinc finger protein, multitype 1 |
380 |
0.82 |
| chr16_87636019_87636210 | 1.07 |
Gm22808 |
predicted gene, 22808 |
14994 |
0.19 |
| chr14_66105986_66106243 | 1.06 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
4546 |
0.18 |
| chr12_104856908_104857124 | 1.05 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
355 |
0.86 |
| chr13_23761212_23761388 | 1.05 |
H4c1 |
H4 clustered histone 1 |
70 |
0.86 |
| chr8_23031563_23031855 | 1.05 |
Ank1 |
ankyrin 1, erythroid |
3390 |
0.23 |
| chr5_66133552_66133721 | 1.03 |
Rbm47 |
RNA binding motif protein 47 |
17320 |
0.11 |
| chr5_32713672_32713870 | 1.02 |
Gm43852 |
predicted gene 43852 |
217 |
0.89 |
| chr14_75242017_75242184 | 1.02 |
Cpb2 |
carboxypeptidase B2 (plasma) |
187 |
0.93 |
| chr14_41167462_41167613 | 1.02 |
Sftpd |
surfactant associated protein D |
15598 |
0.1 |
| chr10_42089474_42089633 | 1.01 |
Tdg-ps2 |
thymine DNA glycosylase, pseudogene 2 |
34247 |
0.18 |
| chr15_80836341_80836511 | 1.01 |
Tnrc6b |
trinucleotide repeat containing 6b |
37711 |
0.15 |
| chr18_16608064_16608271 | 1.01 |
Cdh2 |
cadherin 2 |
61902 |
0.14 |
| chr4_89311006_89311177 | 1.00 |
Cdkn2b |
cyclin dependent kinase inhibitor 2B |
52 |
0.97 |
| chr11_6528635_6528936 | 0.99 |
Snhg15 |
small nucleolar RNA host gene 15 |
6 |
0.9 |
| chr10_59476612_59476997 | 0.99 |
Mcu |
mitochondrial calcium uniporter |
18644 |
0.18 |
| chr12_72761006_72761183 | 0.99 |
Ppm1a |
protein phosphatase 1A, magnesium dependent, alpha isoform |
117 |
0.97 |
| chr1_132155864_132156019 | 0.99 |
Cdk18 |
cyclin-dependent kinase 18 |
16257 |
0.11 |
| chr1_74340683_74340845 | 0.99 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
8101 |
0.09 |
| chr5_91139460_91139622 | 0.97 |
Areg |
amphiregulin |
58 |
0.98 |
| chr3_65666168_65666329 | 0.96 |
Lekr1 |
leucine, glutamate and lysine rich 1 |
2 |
0.97 |
| chr3_58414552_58414712 | 0.96 |
Tsc22d2 |
TSC22 domain family, member 2 |
83 |
0.97 |
| chr9_121001064_121001215 | 0.96 |
Ulk4 |
unc-51-like kinase 4 |
40632 |
0.1 |
| chr10_20952125_20952679 | 0.96 |
Ahi1 |
Abelson helper integration site 1 |
145 |
0.97 |
| chr6_113644583_113645202 | 0.95 |
Gm43964 |
predicted gene, 43964 |
3371 |
0.09 |
| chr7_139388680_139389161 | 0.95 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
189 |
0.96 |
| chr7_99965224_99965392 | 0.95 |
Rnf169 |
ring finger protein 169 |
426 |
0.77 |
| chr8_79711664_79711818 | 0.94 |
Abce1 |
ATP-binding cassette, sub-family E (OABP), member 1 |
1 |
0.55 |
| chr13_64152199_64152545 | 0.93 |
Zfp367 |
zinc finger protein 367 |
113 |
0.63 |
| chr9_20492670_20492836 | 0.93 |
Zfp426 |
zinc finger protein 426 |
7 |
0.96 |
| chr7_126898253_126898420 | 0.93 |
Tmem219 |
transmembrane protein 219 |
55 |
0.92 |
| chr5_121191432_121191739 | 0.92 |
Ptpn11 |
protein tyrosine phosphatase, non-receptor type 11 |
188 |
0.92 |
| chr15_85806972_85807337 | 0.92 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
3936 |
0.16 |
| chr15_68363065_68363728 | 0.92 |
Gm20732 |
predicted gene 20732 |
220 |
0.9 |
| chr13_9930031_9930188 | 0.92 |
Gm47406 |
predicted gene, 47406 |
37199 |
0.15 |
| chr17_6828357_6828615 | 0.92 |
4933426B08Rik |
RIKEN cDNA 4933426B08 gene |
19977 |
0.14 |
| chr7_80324131_80324289 | 0.91 |
Rccd1 |
RCC1 domain containing 1 |
57 |
0.95 |
| chr9_74896463_74896757 | 0.91 |
Onecut1 |
one cut domain, family member 1 |
30126 |
0.13 |
| chr4_108425790_108426145 | 0.91 |
Gpx7 |
glutathione peroxidase 7 |
19006 |
0.1 |
| chr4_126677507_126677663 | 0.91 |
Psmb2 |
proteasome (prosome, macropain) subunit, beta type 2 |
45 |
0.97 |
| chr18_65052807_65053179 | 0.90 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
2003 |
0.39 |
| chr16_33966968_33967146 | 0.90 |
Umps |
uridine monophosphate synthetase |
19 |
0.94 |
| chr15_102103356_102103597 | 0.90 |
Tns2 |
tensin 2 |
488 |
0.69 |
| chr3_97226229_97226380 | 0.89 |
Bcl9 |
B cell CLL/lymphoma 9 |
1060 |
0.5 |
| chr5_53995353_53995770 | 0.89 |
Stim2 |
stromal interaction molecule 2 |
2938 |
0.33 |
| chr4_116562088_116562258 | 0.89 |
Gpbp1l1 |
GC-rich promoter binding protein 1-like 1 |
4094 |
0.13 |
| chr5_120566729_120566934 | 0.89 |
2510016D11Rik |
RIKEN cDNA 2510016D11 gene |
13792 |
0.09 |
| chr7_19629444_19629613 | 0.89 |
Relb |
avian reticuloendotheliosis viral (v-rel) oncogene related B |
90 |
0.93 |
| chr11_97619625_97619776 | 0.89 |
2410003L11Rik |
RIKEN cDNA 2410003L11 gene |
4718 |
0.11 |
| chr7_27196105_27196414 | 0.89 |
Snrpa |
small nuclear ribonucleoprotein polypeptide A |
12 |
0.86 |
| chr11_98588166_98588531 | 0.89 |
Ormdl3 |
ORM1-like 3 (S. cerevisiae) |
980 |
0.38 |
| chr2_6165912_6166067 | 0.89 |
A230108P19Rik |
RIKEN cDNA A230108P19 gene |
27262 |
0.12 |
| chr10_75783826_75784124 | 0.88 |
Gstt3 |
glutathione S-transferase, theta 3 |
2561 |
0.13 |
| chr2_36136541_36136723 | 0.88 |
Mrrf |
mitochondrial ribosome recycling factor |
2 |
0.49 |
| chr12_91846606_91846757 | 0.88 |
Sel1l |
sel-1 suppressor of lin-12-like (C. elegans) |
2448 |
0.23 |
| chr15_55072105_55072274 | 0.88 |
Taf2 |
TATA-box binding protein associated factor 2 |
37 |
0.97 |
| chr5_113142295_113142468 | 0.87 |
4930557B06Rik |
RIKEN cDNA 4930557B06 gene |
68 |
0.95 |
| chr10_128547392_128547567 | 0.87 |
Zc3h10 |
zinc finger CCCH type containing 10 |
160 |
0.84 |
| chr7_118704359_118704549 | 0.87 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
926 |
0.45 |
| chr2_20896513_20896664 | 0.87 |
Arhgap21 |
Rho GTPase activating protein 21 |
13359 |
0.21 |
| chr9_30820894_30821215 | 0.86 |
Gm31013 |
predicted gene, 31013 |
39575 |
0.16 |
| chr13_64181816_64182068 | 0.86 |
Habp4 |
hyaluronic acid binding protein 4 |
2561 |
0.17 |
| chr10_67111164_67111328 | 0.86 |
Reep3 |
receptor accessory protein 3 |
14301 |
0.19 |
| chr8_40881703_40881854 | 0.85 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
6830 |
0.17 |
| chr10_21144969_21145120 | 0.85 |
Gm26577 |
predicted gene, 26577 |
50 |
0.96 |
| chr14_69411185_69411411 | 0.84 |
Gm38378 |
predicted gene, 38378 |
13527 |
0.11 |
| chr1_165236971_165237140 | 0.84 |
Tiprl |
TIP41, TOR signalling pathway regulator-like (S. cerevisiae) |
59 |
0.96 |
| chr11_102742573_102742724 | 0.84 |
Gm16342 |
predicted gene 16342 |
10243 |
0.11 |
| chr16_38562717_38562882 | 0.83 |
Tmem39a |
transmembrane protein 39a |
13 |
0.97 |
| chr6_145855716_145856207 | 0.83 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr5_125484345_125484496 | 0.82 |
Gm27551 |
predicted gene, 27551 |
5043 |
0.14 |
| chr2_153161441_153161599 | 0.82 |
Tm9sf4 |
transmembrane 9 superfamily protein member 4 |
21 |
0.97 |
| chr4_11966482_11966677 | 0.82 |
1700123M08Rik |
RIKEN cDNA 1700123M08 gene |
5 |
0.62 |
| chr3_89458992_89459146 | 0.82 |
Pmvk |
phosphomevalonate kinase |
49 |
0.95 |
| chr15_28181897_28182071 | 0.81 |
Gm49234 |
predicted gene, 49234 |
10138 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 1.4 | 9.9 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.8 | 4.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.5 | 1.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.5 | 1.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 | 1.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.4 | 0.9 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.4 | 2.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 1.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 | 1.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.4 | 1.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 5.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.3 | 2.3 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.3 | 1.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 0.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 0.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 0.9 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 1.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 0.8 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 0.8 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 | 1.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 2.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.2 | 0.7 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 1.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 | 2.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 0.7 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.2 | 2.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 1.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 2.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 1.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 0.4 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.2 | 0.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 0.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.6 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 0.5 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.2 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 0.7 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 1.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 0.3 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.2 | 0.5 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.5 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 1.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 1.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 1.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.4 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.1 | 0.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.7 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.1 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.0 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.5 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 0.4 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 0.3 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.8 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 0.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.4 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 | 0.7 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.1 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.4 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.4 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 1.5 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 0.2 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.9 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.1 | 0.4 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.8 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 1.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.3 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.7 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 2.0 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 1.0 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.1 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.7 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.1 | 1.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.7 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 1.2 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.5 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.8 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.5 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.2 | GO:0060177 | regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 1.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.2 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.3 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 4.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 3.2 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.5 | GO:0006582 | melanin metabolic process(GO:0006582) |
| 0.0 | 0.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.2 | GO:0072160 | nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.0 | 0.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.3 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.5 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.2 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 | 0.4 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.2 | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching(GO:0045829) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.0 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.0 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.9 | GO:0034341 | response to interferon-gamma(GO:0034341) |
| 0.0 | 0.1 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 0.0 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 1.6 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.0 | 0.1 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.0 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.0 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.0 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 4.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.4 | 1.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.4 | 1.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 1.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 2.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 3.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.8 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 2.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 1.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.3 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 1.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 1.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 0.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 1.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.2 | GO:0018854 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 4.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.6 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 9.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 1.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.5 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 2.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.1 | GO:0018423 | protein C-terminal leucine carboxyl O-methyltransferase activity(GO:0018423) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.3 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 1.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0004337 | geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 6.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 2.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0018733 | 3,4-dihydrocoumarin hydrolase activity(GO:0018733) |
| 0.0 | 1.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.4 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.0 | GO:0030352 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0030580 | C-methyltransferase activity(GO:0008169) 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.0 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 8.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 2.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.4 | 10.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 4.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 4.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |