Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.96
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
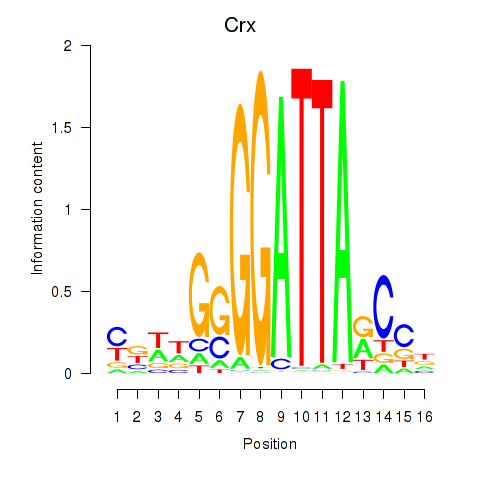
Crx
|
ENSMUSG00000041578.9 | cone-rod homeobox |
|
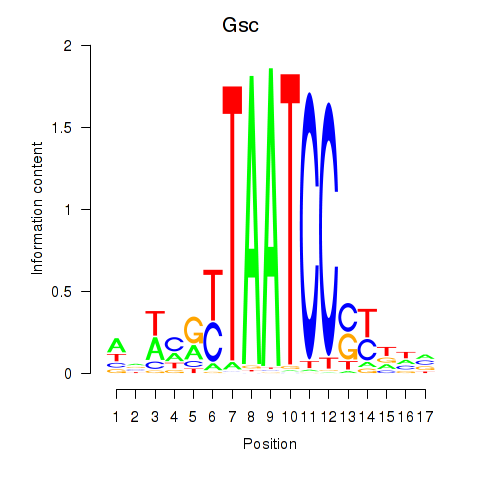
Gsc
|
ENSMUSG00000021095.4 | goosecoid homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_104448270_104448434 | Gsc | 24978 | 0.112130 | -0.52 | 2.9e-01 | Click! |
| chr12_104467528_104467691 | Gsc | 5721 | 0.154405 | 0.40 | 4.3e-01 | Click! |
| chr12_104454871_104455035 | Gsc | 18377 | 0.125087 | -0.10 | 8.5e-01 | Click! |
Activity of the Crx_Gsc motif across conditions
Conditions sorted by the z-value of the Crx_Gsc motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
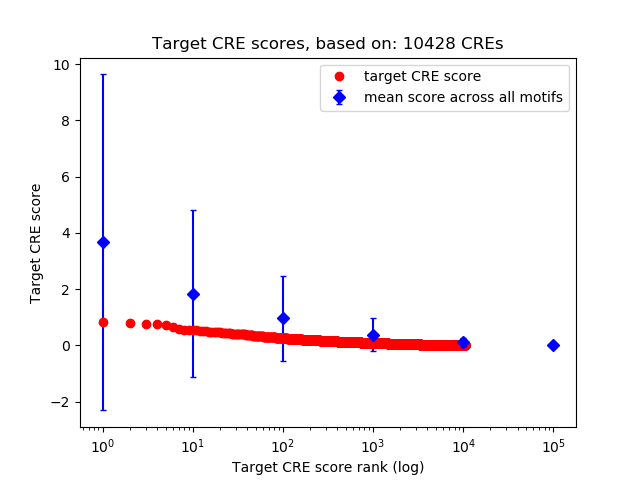
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_19452656_19452840 | 0.84 |
Gm33104 |
predicted gene, 33104 |
33967 |
0.17 |
| chr17_63010676_63010827 | 0.81 |
Gm25348 |
predicted gene, 25348 |
86249 |
0.1 |
| chr11_16854254_16854692 | 0.76 |
Egfr |
epidermal growth factor receptor |
23677 |
0.17 |
| chr3_62404931_62405082 | 0.76 |
Arhgef26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
14662 |
0.25 |
| chr12_104343910_104344587 | 0.73 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
5762 |
0.12 |
| chr7_4815487_4815639 | 0.66 |
Ube2s |
ubiquitin-conjugating enzyme E2S |
2973 |
0.11 |
| chr3_89147086_89147561 | 0.57 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
3891 |
0.08 |
| chr19_18145861_18146289 | 0.56 |
Gm18610 |
predicted gene, 18610 |
59720 |
0.14 |
| chr17_57206582_57206733 | 0.56 |
Mir6978 |
microRNA 6978 |
10571 |
0.1 |
| chr6_51720326_51720477 | 0.55 |
Gm38811 |
predicted gene, 38811 |
9320 |
0.26 |
| chr11_16906555_16906817 | 0.55 |
Egfr |
epidermal growth factor receptor |
1501 |
0.41 |
| chr11_16873480_16873631 | 0.52 |
Egfr |
epidermal growth factor receptor |
4595 |
0.23 |
| chr9_74791375_74791538 | 0.51 |
Gm22315 |
predicted gene, 22315 |
9386 |
0.19 |
| chr19_8835906_8836201 | 0.49 |
Bscl2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
1414 |
0.13 |
| chr11_16901525_16901732 | 0.48 |
Egfr |
epidermal growth factor receptor |
3557 |
0.24 |
| chr11_5909584_5909735 | 0.48 |
Gck |
glucokinase |
5465 |
0.12 |
| chr9_106236583_106237078 | 0.47 |
Alas1 |
aminolevulinic acid synthase 1 |
254 |
0.85 |
| chr12_87443142_87443299 | 0.47 |
Alkbh1 |
alkB homolog 1, histone H2A dioxygenase |
606 |
0.34 |
| chr1_88048809_88049389 | 0.46 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6289 |
0.08 |
| chr17_64643456_64643864 | 0.46 |
Man2a1 |
mannosidase 2, alpha 1 |
42924 |
0.17 |
| chr18_81199291_81199673 | 0.45 |
4930594M17Rik |
RIKEN cDNA 4930594M17 gene |
36584 |
0.17 |
| chr4_63229865_63230144 | 0.44 |
Col27a1 |
collagen, type XXVII, alpha 1 |
4282 |
0.2 |
| chr2_134489871_134490022 | 0.44 |
Hao1 |
hydroxyacid oxidase 1, liver |
64361 |
0.14 |
| chr15_7201530_7201687 | 0.44 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
21461 |
0.23 |
| chr8_40657023_40657174 | 0.43 |
Adam24 |
a disintegrin and metallopeptidase domain 24 (testase 1) |
17979 |
0.13 |
| chr8_125349423_125349587 | 0.42 |
Gm16237 |
predicted gene 16237 |
122182 |
0.05 |
| chr2_72207332_72207483 | 0.42 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
8608 |
0.18 |
| chr15_7189723_7189941 | 0.41 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
33237 |
0.19 |
| chr6_86699596_86699790 | 0.41 |
Gm44214 |
predicted gene, 44214 |
11265 |
0.1 |
| chr1_88064051_88064407 | 0.41 |
AC087801.2 |
UDP glucuronosyltransferase 1 family, polypeptide A9 (Ugt1a9), pseudogene |
3105 |
0.1 |
| chr19_31884765_31884939 | 0.40 |
A1cf |
APOBEC1 complementation factor |
16071 |
0.21 |
| chr5_23562685_23562882 | 0.40 |
Srpk2 |
serine/arginine-rich protein specific kinase 2 |
13413 |
0.19 |
| chr12_35681929_35682122 | 0.40 |
9130015A21Rik |
RIKEN cDNA 9130015A21 gene |
4279 |
0.24 |
| chr1_168682011_168682173 | 0.40 |
1700063I16Rik |
RIKEN cDNA 1700063I16 gene |
6210 |
0.33 |
| chr9_74672657_74672818 | 0.40 |
Gm27233 |
predicted gene 27233 |
36525 |
0.18 |
| chr17_86552135_86552300 | 0.40 |
Gm10309 |
predicted gene 10309 |
46985 |
0.15 |
| chr11_16782319_16782716 | 0.38 |
Egfr |
epidermal growth factor receptor |
30287 |
0.16 |
| chr11_16849295_16849477 | 0.38 |
Egfros |
epidermal growth factor receptor, opposite strand |
18684 |
0.19 |
| chr9_107649565_107649816 | 0.38 |
Slc38a3 |
solute carrier family 38, member 3 |
6330 |
0.08 |
| chr9_74866346_74866537 | 0.38 |
Onecut1 |
one cut domain, family member 1 |
43 |
0.97 |
| chr13_41386761_41386912 | 0.37 |
Gm48571 |
predicted gene, 48571 |
6880 |
0.15 |
| chr15_95039554_95039720 | 0.37 |
Gm23129 |
predicted gene, 23129 |
52499 |
0.17 |
| chr6_108667181_108667428 | 0.37 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
4258 |
0.19 |
| chr11_16589661_16589834 | 0.36 |
Gm12663 |
predicted gene 12663 |
46319 |
0.12 |
| chr6_70874727_70874899 | 0.36 |
Eif2ak3 |
eukaryotic translation initiation factor 2 alpha kinase 3 |
3748 |
0.18 |
| chr11_65731180_65731331 | 0.36 |
Mir744 |
microRNA 744 |
3577 |
0.27 |
| chr10_95354659_95354831 | 0.35 |
2310039L15Rik |
RIKEN cDNA 2310039L15 gene |
143 |
0.94 |
| chr1_36228499_36228653 | 0.34 |
Uggt1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
15531 |
0.16 |
| chr19_12700970_12701121 | 0.34 |
Keg1 |
kidney expressed gene 1 |
5231 |
0.11 |
| chr1_88040499_88040687 | 0.34 |
AC087801.1 |
UDP glycosyltransferase 1 family (Ytg1) pseudogene |
2993 |
0.1 |
| chr11_16850518_16850669 | 0.34 |
Egfros |
epidermal growth factor receptor, opposite strand |
19891 |
0.18 |
| chr14_71251214_71251453 | 0.34 |
Gm4251 |
predicted gene 4251 |
49 |
0.99 |
| chr2_72150298_72150627 | 0.33 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
1666 |
0.39 |
| chr7_97424078_97424348 | 0.32 |
Thrsp |
thyroid hormone responsive |
6483 |
0.13 |
| chr11_94239763_94239948 | 0.32 |
Wfikkn2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
2852 |
0.23 |
| chr7_140769076_140769314 | 0.32 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
4714 |
0.1 |
| chr8_114155044_114155305 | 0.32 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
21532 |
0.24 |
| chr13_112283095_112283277 | 0.32 |
Ankrd55 |
ankyrin repeat domain 55 |
5265 |
0.19 |
| chr12_32681759_32681933 | 0.32 |
Gm47937 |
predicted gene, 47937 |
8552 |
0.23 |
| chr3_30802440_30802630 | 0.31 |
4933429H19Rik |
RIKEN cDNA 4933429H19 gene |
8971 |
0.14 |
| chr18_20057458_20057675 | 0.31 |
Dsc2 |
desmocollin 2 |
1912 |
0.41 |
| chr11_16780614_16780813 | 0.30 |
Egfr |
epidermal growth factor receptor |
28483 |
0.16 |
| chr4_102646817_102646971 | 0.30 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
57051 |
0.14 |
| chr19_53781327_53781513 | 0.30 |
Rbm20 |
RNA binding motif protein 20 |
11888 |
0.17 |
| chr14_18221191_18221360 | 0.30 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
8903 |
0.14 |
| chr12_40218503_40218731 | 0.30 |
Ifrd1 |
interferon-related developmental regulator 1 |
1104 |
0.33 |
| chr1_41557234_41557385 | 0.30 |
Gm28634 |
predicted gene 28634 |
27766 |
0.27 |
| chr8_76540217_76540646 | 0.30 |
Gm27355 |
predicted gene, 27355 |
78851 |
0.1 |
| chr6_149194053_149194312 | 0.30 |
Amn1 |
antagonist of mitotic exit network 1 |
5470 |
0.16 |
| chrX_48054302_48054477 | 0.30 |
Apln |
apelin |
19536 |
0.2 |
| chr5_102008790_102008983 | 0.29 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
27328 |
0.16 |
| chr7_97414019_97414355 | 0.29 |
Thrsp |
thyroid hormone responsive |
3332 |
0.16 |
| chr10_79350263_79350435 | 0.29 |
Vmn2r82 |
vomeronasal 2, receptor 82 |
6171 |
0.22 |
| chr7_97569323_97569597 | 0.29 |
Aamdc |
adipogenesis associated Mth938 domain containing |
6193 |
0.13 |
| chr3_69280240_69280391 | 0.29 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
36546 |
0.13 |
| chr1_174842692_174842843 | 0.29 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
79052 |
0.11 |
| chr8_77009522_77009673 | 0.29 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
101325 |
0.07 |
| chr11_44520601_44520776 | 0.28 |
Rnf145 |
ring finger protein 145 |
383 |
0.85 |
| chr15_67162617_67162836 | 0.28 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
13986 |
0.25 |
| chr7_70346773_70346973 | 0.28 |
Gm44948 |
predicted gene 44948 |
823 |
0.39 |
| chr9_57679444_57679669 | 0.28 |
Cyp1a2 |
cytochrome P450, family 1, subfamily a, polypeptide 2 |
2480 |
0.18 |
| chr16_81387153_81387317 | 0.28 |
Ncam2 |
neural cell adhesion molecule 2 |
78388 |
0.11 |
| chr17_37190817_37190981 | 0.28 |
Ubd |
ubiquitin D |
2993 |
0.09 |
| chr1_84758953_84759122 | 0.27 |
Trip12 |
thyroid hormone receptor interactor 12 |
1147 |
0.47 |
| chr18_36656472_36656632 | 0.27 |
Ankhd1 |
ankyrin repeat and KH domain containing 1 |
1042 |
0.31 |
| chr6_119362732_119362913 | 0.27 |
Adipor2 |
adiponectin receptor 2 |
25864 |
0.16 |
| chr1_88044946_88045337 | 0.27 |
AC087801.1 |
UDP glycosyltransferase 1 family (Ytg1) pseudogene |
7541 |
0.08 |
| chr18_61675552_61675912 | 0.27 |
Il17b |
interleukin 17B |
9155 |
0.1 |
| chr2_142804438_142804612 | 0.27 |
Gm22310 |
predicted gene, 22310 |
17629 |
0.25 |
| chr11_118254084_118254269 | 0.27 |
Cyth1 |
cytohesin 1 |
5584 |
0.17 |
| chr18_75498893_75499229 | 0.27 |
Gm10532 |
predicted gene 10532 |
15584 |
0.25 |
| chr19_46741577_46741728 | 0.27 |
Cnnm2 |
cyclin M2 |
19944 |
0.14 |
| chr15_58902616_58902767 | 0.27 |
Rnf139 |
ring finger protein 139 |
2554 |
0.18 |
| chr2_118756278_118756444 | 0.26 |
Ccdc9b |
coiled-coil domain containing 9B |
6264 |
0.12 |
| chr5_66114502_66114794 | 0.26 |
Rbm47 |
RNA binding motif protein 47 |
16457 |
0.11 |
| chr14_30566208_30566359 | 0.26 |
Tkt |
transketolase |
1716 |
0.29 |
| chr11_16892807_16893180 | 0.26 |
Egfr |
epidermal growth factor receptor |
12192 |
0.19 |
| chrX_168899245_168899407 | 0.26 |
Arhgap6 |
Rho GTPase activating protein 6 |
104227 |
0.07 |
| chr4_49566076_49566232 | 0.26 |
Aldob |
aldolase B, fructose-bisphosphate |
16608 |
0.13 |
| chr14_57107820_57108112 | 0.26 |
Gjb2 |
gap junction protein, beta 2 |
3264 |
0.19 |
| chr9_122849425_122849576 | 0.26 |
Gm47140 |
predicted gene, 47140 |
1082 |
0.34 |
| chr6_128035036_128035224 | 0.26 |
Tspan9 |
tetraspanin 9 |
125 |
0.96 |
| chr13_112271987_112272144 | 0.26 |
Ankrd55 |
ankyrin repeat domain 55 |
16386 |
0.16 |
| chr1_34389167_34389350 | 0.26 |
Gm5266 |
predicted gene 5266 |
21887 |
0.09 |
| chr8_125133982_125134146 | 0.25 |
Disc1 |
disrupted in schizophrenia 1 |
46077 |
0.14 |
| chr14_63273010_63273187 | 0.25 |
Gata4 |
GATA binding protein 4 |
1406 |
0.4 |
| chr15_77153006_77153219 | 0.25 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
444 |
0.78 |
| chr1_132362082_132362267 | 0.25 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
2774 |
0.18 |
| chr13_44409176_44409378 | 0.25 |
Gm33958 |
predicted gene, 33958 |
18160 |
0.15 |
| chr19_10097022_10097275 | 0.24 |
Fads2 |
fatty acid desaturase 2 |
4598 |
0.16 |
| chr2_134827983_134828240 | 0.24 |
Gm14036 |
predicted gene 14036 |
24162 |
0.2 |
| chr16_87610392_87610586 | 0.24 |
Gm22808 |
predicted gene, 22808 |
10631 |
0.19 |
| chr11_84866770_84866933 | 0.24 |
Ggnbp2 |
gametogenetin binding protein 2 |
3351 |
0.15 |
| chr7_144941851_144942303 | 0.24 |
Ccnd1 |
cyclin D1 |
2152 |
0.23 |
| chr17_10745264_10745425 | 0.24 |
Gm16169 |
predicted gene 16169 |
62490 |
0.13 |
| chr4_6261154_6261352 | 0.24 |
Gm11798 |
predicted gene 11798 |
288 |
0.91 |
| chr5_65390468_65390702 | 0.24 |
Rpl9 |
ribosomal protein L9 |
248 |
0.79 |
| chr5_120480072_120480223 | 0.24 |
Sds |
serine dehydratase |
216 |
0.8 |
| chr13_111287126_111287324 | 0.24 |
Actbl2 |
actin, beta-like 2 |
32212 |
0.16 |
| chr5_90314911_90315062 | 0.23 |
Ankrd17 |
ankyrin repeat domain 17 |
10882 |
0.21 |
| chr12_108856815_108857551 | 0.23 |
Gm22079 |
predicted gene, 22079 |
5486 |
0.1 |
| chr17_12168642_12168825 | 0.23 |
Agpat4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
16961 |
0.18 |
| chr9_122863893_122864050 | 0.23 |
Zfp445 |
zinc finger protein 445 |
1627 |
0.22 |
| chr11_107337882_107338046 | 0.23 |
Gm11716 |
predicted gene 11716 |
86 |
0.79 |
| chr8_125381516_125381735 | 0.23 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
111085 |
0.06 |
| chr4_53153864_53154038 | 0.23 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
5944 |
0.24 |
| chr19_12654451_12654602 | 0.23 |
Gm24521 |
predicted gene, 24521 |
10729 |
0.09 |
| chr5_96979772_96979923 | 0.23 |
Gm9484 |
predicted gene 9484 |
17517 |
0.11 |
| chr3_116955207_116955371 | 0.23 |
Gm42892 |
predicted gene 42892 |
10959 |
0.13 |
| chr10_86350127_86350278 | 0.22 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
47348 |
0.13 |
| chr11_110380321_110380711 | 0.22 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
18606 |
0.23 |
| chr7_140768834_140769022 | 0.22 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
4447 |
0.1 |
| chr4_107999051_107999218 | 0.22 |
Slc1a7 |
solute carrier family 1 (glutamate transporter), member 7 |
10883 |
0.13 |
| chr5_66559778_66559933 | 0.22 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
12898 |
0.15 |
| chr9_122848440_122848788 | 0.22 |
Gm47140 |
predicted gene, 47140 |
196 |
0.89 |
| chr2_52575707_52576080 | 0.22 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
17326 |
0.18 |
| chr1_9981778_9981956 | 0.22 |
Gm15818 |
predicted gene 15818 |
520 |
0.62 |
| chr19_42678230_42678434 | 0.22 |
Gm25216 |
predicted gene, 25216 |
21570 |
0.17 |
| chr6_93348262_93348414 | 0.22 |
Gm25094 |
predicted gene, 25094 |
12750 |
0.26 |
| chr12_101875225_101875382 | 0.22 |
Trip11 |
thyroid hormone receptor interactor 11 |
8304 |
0.15 |
| chr4_46139069_46139220 | 0.22 |
Tstd2 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
450 |
0.51 |
| chr7_39524202_39524356 | 0.22 |
Zfp619 |
zinc finger protein 619 |
6513 |
0.13 |
| chr5_54622556_54622707 | 0.22 |
Gm8069 |
predicted pseudogene 8069 |
29490 |
0.22 |
| chr17_12895514_12895853 | 0.22 |
Pnldc1 |
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
99 |
0.91 |
| chr8_9500530_9500773 | 0.22 |
4930435N07Rik |
RIKEN cDNA 4930435N07 gene |
41559 |
0.13 |
| chr15_93825347_93825644 | 0.22 |
Gm49445 |
predicted gene, 49445 |
457 |
0.87 |
| chr3_122890081_122890252 | 0.22 |
Fabp2 |
fatty acid binding protein 2, intestinal |
4906 |
0.14 |
| chr16_26678560_26678733 | 0.21 |
Il1rap |
interleukin 1 receptor accessory protein |
43788 |
0.18 |
| chr8_61436826_61436991 | 0.21 |
Cbr4 |
carbonyl reductase 4 |
50826 |
0.12 |
| chr2_68932763_68932937 | 0.21 |
Cers6 |
ceramide synthase 6 |
14410 |
0.14 |
| chr1_10156105_10156276 | 0.21 |
Arfgef1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) |
1660 |
0.36 |
| chr1_71807937_71808302 | 0.21 |
Gm37217 |
predicted gene, 37217 |
38491 |
0.15 |
| chr15_91192098_91192262 | 0.21 |
Abcd2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
381 |
0.87 |
| chr11_51598115_51598266 | 0.21 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
1077 |
0.39 |
| chr11_70057900_70058083 | 0.21 |
Asgr1 |
asialoglycoprotein receptor 1 |
2110 |
0.15 |
| chr5_63953741_63953922 | 0.21 |
Rell1 |
RELT-like 1 |
14992 |
0.15 |
| chr13_31401311_31401475 | 0.21 |
G630018N14Rik |
RIKEN cDNA G630018N14 gene |
7529 |
0.16 |
| chr7_19560953_19561121 | 0.21 |
Ppp1r37 |
protein phosphatase 1, regulatory subunit 37 |
2039 |
0.16 |
| chr14_120313311_120313499 | 0.21 |
Mbnl2 |
muscleblind like splicing factor 2 |
6949 |
0.28 |
| chr1_73630126_73630279 | 0.21 |
C530043A13Rik |
RIKEN cDNA C530043A13 gene |
100412 |
0.06 |
| chr19_26765890_26766041 | 0.21 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
4526 |
0.25 |
| chr13_74506069_74506220 | 0.21 |
Gm49763 |
predicted gene, 49763 |
12001 |
0.1 |
| chr4_102576883_102577034 | 0.21 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
6863 |
0.31 |
| chr1_174900896_174901097 | 0.21 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
20823 |
0.27 |
| chr9_57686463_57686734 | 0.21 |
Cyp1a1 |
cytochrome P450, family 1, subfamily a, polypeptide 1 |
1330 |
0.3 |
| chr7_67609230_67609410 | 0.20 |
Lrrc28 |
leucine rich repeat containing 28 |
2058 |
0.25 |
| chr5_28374728_28374917 | 0.20 |
Rbm33 |
RNA binding motif protein 33 |
19233 |
0.22 |
| chr12_104081605_104081762 | 0.20 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
1034 |
0.36 |
| chr2_134524166_134524489 | 0.20 |
Hao1 |
hydroxyacid oxidase 1, liver |
29980 |
0.24 |
| chr1_54496220_54496429 | 0.20 |
Gm36955 |
predicted gene, 36955 |
23946 |
0.15 |
| chr6_67385240_67385404 | 0.20 |
Gm8574 |
predicted gene 8574 |
19 |
0.97 |
| chr5_64075656_64075807 | 0.20 |
Pgm2 |
phosphoglucomutase 2 |
17219 |
0.12 |
| chr2_31745875_31746037 | 0.20 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
13987 |
0.14 |
| chr15_59061763_59061914 | 0.20 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6374 |
0.24 |
| chr8_66587441_66587788 | 0.20 |
Gm16330 |
predicted gene 16330 |
1394 |
0.48 |
| chr19_60532004_60532219 | 0.20 |
Cacul1 |
CDK2 associated, cullin domain 1 |
5438 |
0.23 |
| chr15_35876710_35876861 | 0.20 |
Vps13b |
vacuolar protein sorting 13B |
5063 |
0.18 |
| chr11_16837434_16837735 | 0.20 |
Egfros |
epidermal growth factor receptor, opposite strand |
6882 |
0.22 |
| chr4_117125610_117126073 | 0.20 |
Btbd19 |
BTB (POZ) domain containing 19 |
116 |
0.86 |
| chr6_48684930_48685081 | 0.20 |
Gimap4 |
GTPase, IMAP family member 4 |
423 |
0.63 |
| chr14_87140699_87141139 | 0.20 |
Diaph3 |
diaphanous related formin 3 |
224 |
0.95 |
| chr13_34898393_34898709 | 0.20 |
Prpf4b |
pre-mRNA processing factor 4B |
3098 |
0.15 |
| chr4_73048259_73048424 | 0.20 |
Gm11251 |
predicted gene 11251 |
18522 |
0.24 |
| chr12_83905337_83905488 | 0.20 |
Numb |
NUMB endocytic adaptor protein |
16321 |
0.11 |
| chr12_57650772_57650931 | 0.20 |
Ttc6 |
tetratricopeptide repeat domain 6 |
1436 |
0.35 |
| chr7_34226333_34226490 | 0.20 |
Gpi1 |
glucose-6-phosphate isomerase 1 |
2586 |
0.14 |
| chr2_27769095_27769412 | 0.20 |
Rxra |
retinoid X receptor alpha |
29052 |
0.21 |
| chr4_3408345_3408692 | 0.20 |
Gm11784 |
predicted gene 11784 |
21672 |
0.18 |
| chr16_9588741_9588894 | 0.19 |
Grin2a |
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
208516 |
0.02 |
| chr8_76151909_76152060 | 0.19 |
Gm45742 |
predicted gene 45742 |
34957 |
0.2 |
| chr18_12948248_12948402 | 0.19 |
Osbpl1a |
oxysterol binding protein-like 1A |
6484 |
0.18 |
| chr5_148978578_148978772 | 0.19 |
Gm42791 |
predicted gene 42791 |
2418 |
0.14 |
| chr4_102566844_102566995 | 0.19 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
3176 |
0.38 |
| chr2_167292312_167292496 | 0.19 |
Gm11473 |
predicted gene 11473 |
10627 |
0.16 |
| chr5_77406298_77406449 | 0.19 |
Igfbp7 |
insulin-like growth factor binding protein 7 |
1667 |
0.3 |
| chr11_16799047_16799198 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
31580 |
0.16 |
| chr15_25942411_25942580 | 0.19 |
Retreg1 |
reticulophagy regulator 1 |
184 |
0.95 |
| chr9_107298322_107298733 | 0.19 |
Cish |
cytokine inducible SH2-containing protein |
628 |
0.48 |
| chr13_62867105_62867323 | 0.19 |
Fbp1 |
fructose bisphosphatase 1 |
5544 |
0.14 |
| chr2_132677140_132677304 | 0.19 |
Gm14098 |
predicted gene 14098 |
1047 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.2 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.1 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.0 | GO:1904833 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0043307 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.0 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0098739 | import across plasma membrane(GO:0098739) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.0 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.0 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.0 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |