Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
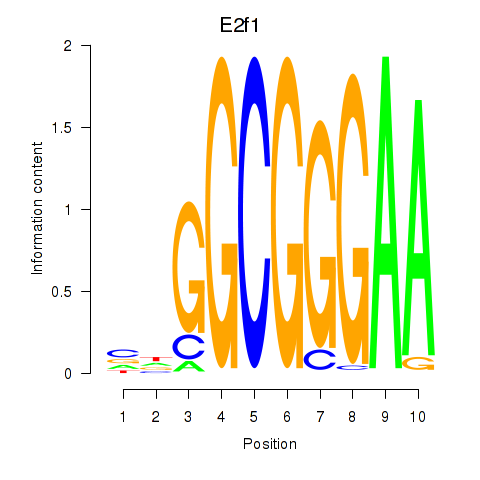
Results for E2f1
Z-value: 1.89
Transcription factors associated with E2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f1
|
ENSMUSG00000027490.11 | E2F transcription factor 1 |
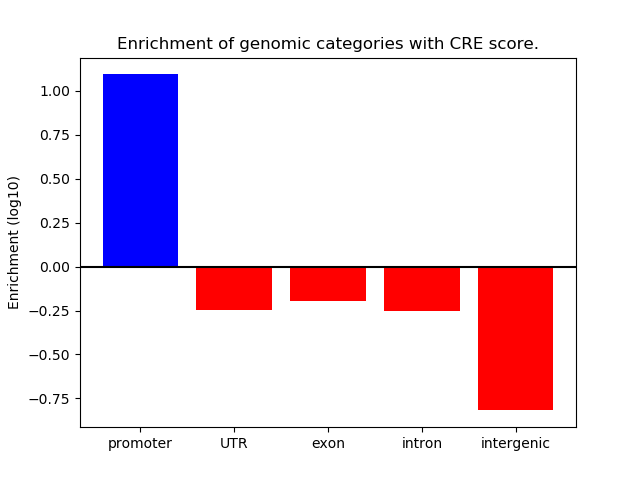
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_154584077_154584353 | E2f1 | 14323 | 0.095175 | 0.84 | 3.5e-02 | Click! |
| chr2_154585868_154586214 | E2f1 | 16149 | 0.092810 | 0.80 | 5.8e-02 | Click! |
| chr2_154570264_154570435 | E2f1 | 457 | 0.695875 | 0.78 | 6.6e-02 | Click! |
| chr2_154585580_154585751 | E2f1 | 15773 | 0.093298 | 0.69 | 1.3e-01 | Click! |
| chr2_154585359_154585533 | E2f1 | 15554 | 0.093582 | 0.55 | 2.6e-01 | Click! |
Activity of the E2f1 motif across conditions
Conditions sorted by the z-value of the E2f1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
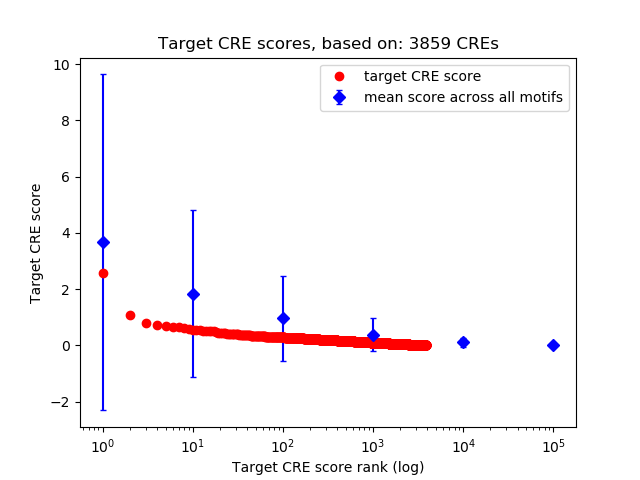
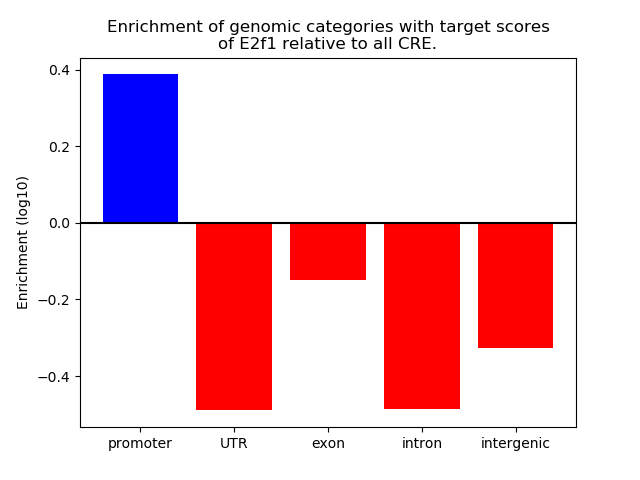
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_28886991_28887442 | 2.58 |
Gm48080 |
predicted gene, 48080 |
834 |
0.6 |
| chr17_32493567_32493723 | 1.07 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
1487 |
0.3 |
| chr16_20621460_20621649 | 0.81 |
Camk2n2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
267 |
0.75 |
| chr9_65333970_65334121 | 0.73 |
Gm39363 |
predicted gene, 39363 |
1525 |
0.2 |
| chr6_47453879_47454048 | 0.68 |
Cul1 |
cullin 1 |
59 |
0.83 |
| chr7_46795972_46796137 | 0.65 |
Hps5 |
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
10 |
0.51 |
| chr1_182495703_182495872 | 0.64 |
Gm37069 |
predicted gene, 37069 |
1631 |
0.28 |
| chr11_4104740_4104947 | 0.60 |
Gm11955 |
predicted gene 11955 |
2912 |
0.12 |
| chr17_28523067_28523337 | 0.57 |
Gm20109 |
predicted gene, 20109 |
54 |
0.53 |
| chr9_110246098_110246279 | 0.56 |
Cspg5 |
chondroitin sulfate proteoglycan 5 |
123 |
0.94 |
| chr6_145767466_145768203 | 0.54 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
17263 |
0.18 |
| chr2_154436614_154436770 | 0.53 |
Cbfa2t2 |
CBFA2/RUNX1 translocation partner 2 |
150 |
0.95 |
| chr8_124751800_124751970 | 0.52 |
Fam89a |
family with sequence similarity 89, member A |
53 |
0.96 |
| chr6_121865645_121865799 | 0.52 |
Mug1 |
murinoglobulin 1 |
19855 |
0.18 |
| chr1_135375014_135375173 | 0.51 |
Shisa4 |
shisa family member 4 |
16 |
0.96 |
| chr1_120111811_120112043 | 0.51 |
Dbi |
diazepam binding inhibitor |
8211 |
0.18 |
| chr13_51992355_51992528 | 0.51 |
Gm26651 |
predicted gene, 26651 |
19227 |
0.2 |
| chr1_136781576_136781746 | 0.46 |
Gm8762 |
predicted gene 8762 |
7859 |
0.14 |
| chr11_119913957_119914303 | 0.44 |
Chmp6 |
charged multivesicular body protein 6 |
115 |
0.95 |
| chr10_40301571_40301722 | 0.44 |
Amd1 |
S-adenosylmethionine decarboxylase 1 |
70 |
0.95 |
| chr7_116039656_116039813 | 0.44 |
1110004F10Rik |
RIKEN cDNA 1110004F10 gene |
22 |
0.95 |
| chr16_10989648_10989826 | 0.43 |
Litaf |
LPS-induced TN factor |
3334 |
0.14 |
| chr1_78511963_78512114 | 0.43 |
BC035947 |
cDNA sequence BC035947 |
120 |
0.57 |
| chr13_98352948_98353139 | 0.42 |
Foxd1 |
forkhead box D1 |
1199 |
0.4 |
| chr2_28840041_28840192 | 0.42 |
Gtf3c4 |
general transcription factor IIIC, polypeptide 4 |
170 |
0.65 |
| chr8_122550942_122551215 | 0.42 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
251 |
0.84 |
| chr17_35861076_35861404 | 0.41 |
Nrm |
nurim (nuclear envelope membrane protein) |
78 |
0.9 |
| chr18_39693461_39693788 | 0.41 |
Gm50398 |
predicted gene, 50398 |
32747 |
0.16 |
| chr6_83473330_83473495 | 0.41 |
Dguok |
deoxyguanosine kinase |
13598 |
0.11 |
| chr16_32670450_32670601 | 0.41 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
1132 |
0.39 |
| chr8_13889986_13890137 | 0.40 |
Coprs |
coordinator of PRMT5, differentiation stimulator |
71 |
0.95 |
| chr12_64918124_64918389 | 0.39 |
Gm527 |
predicted gene 527 |
287 |
0.89 |
| chr10_61719422_61719586 | 0.37 |
Aifm2 |
apoptosis-inducing factor, mitochondrion-associated 2 |
1145 |
0.39 |
| chr1_133508090_133508255 | 0.37 |
Gm8596 |
predicted gene 8596 |
21307 |
0.18 |
| chr13_47105847_47106030 | 0.37 |
Dek |
DEK oncogene (DNA binding) |
108 |
0.84 |
| chr3_90465954_90466142 | 0.37 |
Npr1 |
natriuretic peptide receptor 1 |
182 |
0.87 |
| chr15_78803027_78803209 | 0.37 |
Card10 |
caspase recruitment domain family, member 10 |
76 |
0.95 |
| chr4_53440696_53440847 | 0.36 |
Slc44a1 |
solute carrier family 44, member 1 |
87 |
0.98 |
| chr1_43827441_43827643 | 0.36 |
Uxs1 |
UDP-glucuronate decarboxylase 1 |
199 |
0.93 |
| chr13_23761471_23761669 | 0.36 |
H4c1 |
H4 clustered histone 1 |
340 |
0.53 |
| chr14_61173133_61173377 | 0.35 |
Sacs |
sacsin |
265 |
0.93 |
| chr7_112208684_112208908 | 0.35 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
17060 |
0.24 |
| chr1_51815131_51815349 | 0.35 |
Myo1b |
myosin IB |
5306 |
0.21 |
| chr11_96075628_96076164 | 0.35 |
Atp5g1 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
226 |
0.86 |
| chr15_76369014_76369269 | 0.35 |
Hgh1 |
HGH1 homolog |
243 |
0.79 |
| chr4_63215885_63216054 | 0.35 |
Col27a1 |
collagen, type XXVII, alpha 1 |
534 |
0.76 |
| chr10_63339095_63339253 | 0.34 |
Sirt1 |
sirtuin 1 |
139 |
0.94 |
| chr9_15357682_15357986 | 0.34 |
Cep295 |
centrosomal protein 295 |
46 |
0.92 |
| chr1_136893539_136893740 | 0.34 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
46944 |
0.13 |
| chr17_63863193_63863441 | 0.34 |
Fer |
fer (fms/fps related) protein kinase |
255 |
0.71 |
| chr10_122986042_122986255 | 0.33 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
317 |
0.58 |
| chr8_84872118_84872282 | 0.33 |
Syce2 |
synaptonemal complex central element protein 2 |
7 |
0.93 |
| chr11_61342814_61342965 | 0.33 |
Slc47a2 |
solute carrier family 47, member 2 |
29 |
0.97 |
| chr14_115040574_115040725 | 0.33 |
Mir17hg |
Mir17 host gene (non-protein coding) |
2230 |
0.14 |
| chr4_45470057_45470208 | 0.33 |
Shb |
src homology 2 domain-containing transforming protein B |
12670 |
0.16 |
| chr11_116110160_116110352 | 0.33 |
Trim47 |
tripartite motif-containing 47 |
10 |
0.95 |
| chr18_74065170_74065346 | 0.32 |
Mapk4 |
mitogen-activated protein kinase 4 |
101 |
0.58 |
| chr5_23787298_23787449 | 0.32 |
Rint1 |
RAD50 interactor 1 |
338 |
0.77 |
| chr6_72788306_72788457 | 0.32 |
Tcf7l1 |
transcription factor 7 like 1 (T cell specific, HMG box) |
217 |
0.93 |
| chr8_84011360_84011540 | 0.32 |
Misp3 |
MISP family member 3 |
39 |
0.66 |
| chr10_67051361_67051540 | 0.32 |
Reep3 |
receptor accessory protein 3 |
14128 |
0.18 |
| chr2_163419076_163419281 | 0.32 |
Oser1 |
oxidative stress responsive serine rich 1 |
34 |
0.91 |
| chr5_125441019_125441180 | 0.31 |
Bri3bp |
Bri3 binding protein |
469 |
0.7 |
| chr1_181475926_181476204 | 0.31 |
Ccdc121 |
coiled-coil domain containing 121 |
35386 |
0.14 |
| chr17_29050362_29051086 | 0.31 |
Gm41556 |
predicted gene, 41556 |
2612 |
0.13 |
| chr9_22467821_22467972 | 0.31 |
Rp9 |
retinitis pigmentosa 9 (human) |
274 |
0.86 |
| chr6_134929015_134929166 | 0.31 |
Lockd |
lncRNA downstream of Cdkn1b |
2 |
0.94 |
| chr18_44381017_44381212 | 0.31 |
Dcp2 |
decapping mRNA 2 |
587 |
0.78 |
| chr12_69372296_69372449 | 0.31 |
Gm9887 |
predicted gene 9887 |
86 |
0.68 |
| chr11_80476499_80476662 | 0.31 |
Cdk5r1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
443 |
0.82 |
| chr1_88491899_88492050 | 0.31 |
Glrp1 |
glutamine repeat protein 1 |
18092 |
0.14 |
| chr1_128359251_128359552 | 0.31 |
Mcm6 |
minichromosome maintenance complex component 6 |
246 |
0.91 |
| chr9_47531410_47531633 | 0.31 |
Cadm1 |
cell adhesion molecule 1 |
1148 |
0.49 |
| chr1_72171187_72171446 | 0.30 |
Mreg |
melanoregulin |
40991 |
0.1 |
| chr18_65247932_65248083 | 0.30 |
Mir122 |
microRNA 122 |
854 |
0.54 |
| chr9_119148405_119148556 | 0.30 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
1583 |
0.25 |
| chr2_52858397_52858587 | 0.30 |
Fmnl2 |
formin-like 2 |
624 |
0.82 |
| chr7_48881274_48881434 | 0.30 |
Gm2788 |
predicted gene 2788 |
54 |
0.74 |
| chr8_64767198_64767355 | 0.30 |
Klhl2 |
kelch-like 2, Mayven |
12305 |
0.16 |
| chr4_135516112_135516333 | 0.30 |
Gm25317 |
predicted gene, 25317 |
3743 |
0.16 |
| chr6_87590696_87590858 | 0.30 |
Prokr1 |
prokineticin receptor 1 |
34 |
0.97 |
| chr2_157007239_157007486 | 0.30 |
Dsn1 |
DSN1 homolog, MIS12 kinetochore complex component |
208 |
0.9 |
| chr4_135420119_135420277 | 0.29 |
Mir700 |
microRNA 700 |
3565 |
0.12 |
| chr1_169919370_169919536 | 0.29 |
Ccdc190 |
coiled-coil domain containing 190 |
9195 |
0.17 |
| chr1_79857847_79858438 | 0.29 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
493 |
0.83 |
| chrX_168795419_168795570 | 0.29 |
Arhgap6 |
Rho GTPase activating protein 6 |
395 |
0.89 |
| chr13_31559718_31559877 | 0.29 |
A530084C06Rik |
RIKEN cDNA A530084C06 gene |
464 |
0.73 |
| chr11_106249671_106250177 | 0.29 |
Ftsj3 |
FtsJ RNA methyltransferase homolog 3 (E. coli) |
774 |
0.42 |
| chr2_167570275_167570599 | 0.29 |
Gm11476 |
predicted gene 11476 |
17123 |
0.11 |
| chr2_132252667_132252827 | 0.29 |
Pcna |
proliferating cell nuclear antigen |
145 |
0.85 |
| chr2_167574964_167575153 | 0.29 |
Gm11475 |
predicted gene 11475 |
16337 |
0.11 |
| chr4_153957077_153957238 | 0.28 |
A430005L14Rik |
RIKEN cDNA A430005L14 gene |
80 |
0.95 |
| chr1_93135406_93135557 | 0.28 |
Agxt |
alanine-glyoxylate aminotransferase |
87 |
0.95 |
| chr12_59219022_59219224 | 0.28 |
Fbxo33 |
F-box protein 33 |
322 |
0.86 |
| chr6_29917136_29917315 | 0.28 |
Strip2 |
striatin interacting protein 2 |
212 |
0.88 |
| chr3_19895212_19895377 | 0.28 |
4632415L05Rik |
RIKEN cDNA 4632415L05 gene |
312 |
0.89 |
| chr17_29056624_29056784 | 0.28 |
Gm41556 |
predicted gene, 41556 |
156 |
0.9 |
| chr5_140701925_140702124 | 0.28 |
Iqce |
IQ motif containing E |
55 |
0.97 |
| chr7_143599568_143599932 | 0.28 |
Cars |
cysteinyl-tRNA synthetase |
69 |
0.82 |
| chr11_5541916_5542088 | 0.28 |
Ccdc117 |
coiled-coil domain containing 117 |
185 |
0.92 |
| chr4_129600776_129601168 | 0.28 |
Tmem234 |
transmembrane protein 234 |
11 |
0.81 |
| chr12_21112057_21112211 | 0.28 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
180 |
0.96 |
| chr5_8622651_8622802 | 0.28 |
Rundc3b |
RUN domain containing 3B |
226 |
0.94 |
| chr9_24499400_24499590 | 0.28 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
2323 |
0.31 |
| chr10_91082443_91082594 | 0.28 |
Apaf1 |
apoptotic peptidase activating factor 1 |
89 |
0.84 |
| chr1_21079589_21079810 | 0.28 |
Gm28287 |
predicted gene 28287 |
415 |
0.51 |
| chr7_30535115_30535276 | 0.27 |
Arhgap33 |
Rho GTPase activating protein 33 |
135 |
0.85 |
| chr13_55329990_55330168 | 0.27 |
Mxd3 |
Max dimerization protein 3 |
256 |
0.85 |
| chr4_133369891_133370073 | 0.27 |
Mir5122 |
microRNA 5122 |
206 |
0.57 |
| chr9_21010755_21010906 | 0.27 |
Icam1 |
intercellular adhesion molecule 1 |
5155 |
0.08 |
| chr8_84893555_84893706 | 0.27 |
Gcdh |
glutaryl-Coenzyme A dehydrogenase |
155 |
0.87 |
| chr13_22006021_22006172 | 0.27 |
Prss16 |
protease, serine 16 (thymus) |
316 |
0.68 |
| chr17_85091006_85091157 | 0.27 |
Camkmt |
calmodulin-lysine N-methyltransferase |
475 |
0.65 |
| chr4_141663419_141663714 | 0.27 |
Plekhm2 |
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
552 |
0.67 |
| chr2_181287544_181287702 | 0.27 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
393 |
0.73 |
| chr2_114099402_114099798 | 0.27 |
Aqr |
aquarius |
19587 |
0.16 |
| chr11_49847095_49847246 | 0.27 |
Mapk9 |
mitogen-activated protein kinase 9 |
277 |
0.9 |
| chr1_179847829_179847985 | 0.27 |
Ahctf1 |
AT hook containing transcription factor 1 |
44227 |
0.13 |
| chr18_60601453_60601636 | 0.27 |
Synpo |
synaptopodin |
1047 |
0.47 |
| chr2_151996379_151996542 | 0.27 |
Slc52a3 |
solute carrier protein family 52, member 3 |
51 |
0.96 |
| chr2_38286777_38287020 | 0.26 |
Dennd1a |
DENN/MADD domain containing 1A |
290 |
0.86 |
| chr13_95618213_95618364 | 0.26 |
F2r |
coagulation factor II (thrombin) receptor |
199 |
0.93 |
| chr6_100670825_100671026 | 0.26 |
Shq1 |
SHQ1 homolog (S. cerevisiae) |
208 |
0.92 |
| chr13_21180068_21180231 | 0.26 |
Trim27 |
tripartite motif-containing 27 |
75 |
0.96 |
| chr15_75862455_75863017 | 0.26 |
Gsdmd |
gasdermin D |
357 |
0.74 |
| chr11_21080920_21081322 | 0.26 |
Peli1 |
pellino 1 |
10170 |
0.2 |
| chr15_98805374_98805536 | 0.26 |
B130046B21Rik |
RIKEN cDNA B130046B21 gene |
1569 |
0.19 |
| chr1_133163770_133164045 | 0.26 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
303 |
0.89 |
| chr12_40596271_40596511 | 0.26 |
Dock4 |
dedicator of cytokinesis 4 |
150055 |
0.04 |
| chr18_60848442_60848618 | 0.26 |
Tcof1 |
treacle ribosome biogenesis factor 1 |
84 |
0.96 |
| chr17_6316978_6317224 | 0.26 |
Dynlt1a |
dynein light chain Tctex-type 1A |
317 |
0.82 |
| chr17_53567606_53567787 | 0.26 |
Kat2b |
K(lysine) acetyltransferase 2B |
725 |
0.62 |
| chr7_100492974_100493189 | 0.26 |
Ucp2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
256 |
0.83 |
| chr13_29945538_29945696 | 0.26 |
Gm11365 |
predicted gene 11365 |
5334 |
0.25 |
| chr12_111759014_111759441 | 0.26 |
Klc1 |
kinesin light chain 1 |
214 |
0.89 |
| chr2_103565693_103566028 | 0.26 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
450 |
0.83 |
| chr7_64048693_64048852 | 0.26 |
Gm45054 |
predicted gene 45054 |
7946 |
0.16 |
| chr11_17953583_17953741 | 0.26 |
Etaa1 |
Ewing tumor-associated antigen 1 |
213 |
0.61 |
| chr18_4956286_4956555 | 0.26 |
Svil |
supervillin |
34694 |
0.22 |
| chr5_110110403_110110732 | 0.25 |
Zfp605 |
zinc finger protein 605 |
216 |
0.85 |
| chr4_136177846_136177997 | 0.25 |
E2f2 |
E2F transcription factor 2 |
2862 |
0.19 |
| chr18_54953610_54953761 | 0.25 |
Zfp608 |
zinc finger protein 608 |
3072 |
0.28 |
| chr1_36535791_36536054 | 0.25 |
Ankrd23 |
ankyrin repeat domain 23 |
183 |
0.87 |
| chr4_34730156_34730312 | 0.25 |
Cfap206 |
cilia and flagella associated protein 206 |
28 |
0.97 |
| chr11_20831246_20831415 | 0.25 |
Lgalsl |
lectin, galactoside binding-like |
274 |
0.72 |
| chr4_148140591_148140767 | 0.25 |
Mad2l2 |
MAD2 mitotic arrest deficient-like 2 |
45 |
0.95 |
| chr15_98092785_98093187 | 0.25 |
Senp1 |
SUMO1/sentrin specific peptidase 1 |
99 |
0.64 |
| chr13_12257493_12257701 | 0.25 |
Mtr |
5-methyltetrahydrofolate-homocysteine methyltransferase |
498 |
0.74 |
| chr19_45805094_45805257 | 0.25 |
Kcnip2 |
Kv channel-interacting protein 2 |
1601 |
0.31 |
| chr11_89999415_89999783 | 0.25 |
Pctp |
phosphatidylcholine transfer protein |
3295 |
0.3 |
| chr2_84744034_84744230 | 0.25 |
Gm19426 |
predicted gene, 19426 |
477 |
0.61 |
| chr3_158036818_158036969 | 0.25 |
Lrrc40 |
leucine rich repeat containing 40 |
33 |
0.72 |
| chr7_81706851_81707046 | 0.25 |
Homer2 |
homer scaffolding protein 2 |
4 |
0.97 |
| chr17_21753956_21754159 | 0.25 |
Zfp229 |
zinc finger protein 229 |
18145 |
0.11 |
| chr11_98294523_98294694 | 0.25 |
Gm20644 |
predicted gene 20644 |
9226 |
0.1 |
| chr4_133241348_133241936 | 0.25 |
Map3k6 |
mitogen-activated protein kinase kinase kinase 6 |
824 |
0.47 |
| chr19_44742201_44742385 | 0.25 |
Gm35610 |
predicted gene, 35610 |
940 |
0.5 |
| chr8_122678212_122678419 | 0.25 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
140 |
0.92 |
| chr17_34586879_34587070 | 0.25 |
Notch4 |
notch 4 |
29 |
0.91 |
| chr3_129531047_129531198 | 0.25 |
Gm35986 |
predicted gene, 35986 |
987 |
0.39 |
| chr13_100744587_100744752 | 0.25 |
Mrps36 |
mitochondrial ribosomal protein S36 |
10 |
0.96 |
| chr9_119357444_119357603 | 0.25 |
Xylb |
xylulokinase homolog (H. influenzae) |
46 |
0.96 |
| chr11_51916701_51916882 | 0.25 |
Gm39822 |
predicted gene, 39822 |
17825 |
0.14 |
| chr7_30360114_30360296 | 0.25 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
2567 |
0.1 |
| chr7_128010040_128010379 | 0.25 |
Gm19366 |
predicted gene, 19366 |
2975 |
0.09 |
| chr4_120825407_120825586 | 0.25 |
Nfyc |
nuclear transcription factor-Y gamma |
188 |
0.91 |
| chr5_107437718_107438041 | 0.24 |
Btbd8 |
BTB (POZ) domain containing 8 |
118 |
0.93 |
| chr19_44563003_44563199 | 0.24 |
Hif1an |
hypoxia-inducible factor 1, alpha subunit inhibitor |
251 |
0.88 |
| chr3_59006560_59006721 | 0.24 |
Med12l |
mediator complex subunit 12-like |
153 |
0.8 |
| chr17_30601575_30601726 | 0.24 |
Glo1 |
glyoxalase 1 |
705 |
0.46 |
| chr7_118518801_118518969 | 0.24 |
Coq7 |
demethyl-Q 7 |
7849 |
0.15 |
| chr8_44937447_44937668 | 0.24 |
Fat1 |
FAT atypical cadherin 1 |
2110 |
0.3 |
| chr8_17664781_17664932 | 0.24 |
Csmd1 |
CUB and Sushi multiple domains 1 |
129270 |
0.06 |
| chr10_30200075_30200299 | 0.24 |
Cenpw |
centromere protein W |
305 |
0.92 |
| chr1_59684305_59684456 | 0.24 |
Nop58 |
NOP58 ribonucleoprotein |
591 |
0.56 |
| chr5_124352244_124352411 | 0.24 |
Cdk2ap1 |
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
94 |
0.95 |
| chr13_117024345_117024496 | 0.24 |
Parp8 |
poly (ADP-ribose) polymerase family, member 8 |
853 |
0.64 |
| chr17_26602801_26603141 | 0.24 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
747 |
0.54 |
| chr1_78657322_78657585 | 0.24 |
Acsl3 |
acyl-CoA synthetase long-chain family member 3 |
372 |
0.62 |
| chr16_17210099_17210269 | 0.24 |
Rimbp3 |
RIMS binding protein 3 |
1581 |
0.21 |
| chr15_84855689_84855840 | 0.24 |
Phf21b |
PHD finger protein 21B |
13 |
0.97 |
| chr8_95807155_95807326 | 0.24 |
Cnot1 |
CCR4-NOT transcription complex, subunit 1 |
141 |
0.65 |
| chr7_143978352_143978545 | 0.24 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
23480 |
0.16 |
| chr4_145246125_145246511 | 0.23 |
Tnfrsf1b |
tumor necrosis factor receptor superfamily, member 1b |
552 |
0.79 |
| chr8_12672095_12672260 | 0.23 |
4931415C17Rik |
RIKEN cDNA 4931415C17 gene |
9 |
0.52 |
| chr15_50361351_50361717 | 0.23 |
Gm49198 |
predicted gene, 49198 |
74636 |
0.13 |
| chr13_73733026_73733394 | 0.23 |
Slc12a7 |
solute carrier family 12, member 7 |
116 |
0.96 |
| chr6_6210752_6211010 | 0.23 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
6237 |
0.25 |
| chr6_47920220_47920375 | 0.23 |
Zfp212 |
Zinc finger protein 212 |
179 |
0.92 |
| chr10_52690737_52690888 | 0.23 |
Slc35f1 |
solute carrier family 35, member F1 |
279 |
0.9 |
| chr19_5038948_5039147 | 0.23 |
B4gat1 |
beta-1,4-glucuronyltransferase 1 |
140 |
0.49 |
| chr1_133045746_133045897 | 0.23 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
154 |
0.94 |
| chr11_107547459_107547712 | 0.23 |
Helz |
helicase with zinc finger domain |
345 |
0.7 |
| chr2_70509001_70509542 | 0.23 |
Erich2os |
glutamate rich 2, opposite strand |
72 |
0.66 |
| chr11_104571330_104571492 | 0.23 |
Gm11663 |
predicted gene 11663 |
146 |
0.92 |
| chr11_3408533_3408935 | 0.23 |
Limk2 |
LIM motif-containing protein kinase 2 |
440 |
0.75 |
| chr7_101741464_101741615 | 0.23 |
Clpb |
ClpB caseinolytic peptidase B |
22040 |
0.13 |
| chr4_114909130_114909304 | 0.23 |
Foxd2os |
forkhead box D2, opposite strand |
40 |
0.83 |
| chr4_143161589_143161992 | 0.23 |
Gm13039 |
predicted gene 13039 |
4813 |
0.16 |
| chr14_55671605_55671777 | 0.23 |
Nedd8 |
neural precursor cell expressed, developmentally down-regulated gene 8 |
182 |
0.53 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.4 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.3 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.2 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.2 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 0.2 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.1 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.3 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.1 | 0.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.1 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.4 | GO:0070202 | regulation of establishment of protein localization to chromosome(GO:0070202) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.4 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.2 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0070666 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.3 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.3 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0006056 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.0 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.0 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation(GO:0003198) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0071351 | response to interleukin-18(GO:0070673) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.0 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.0 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:1902592 | viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0030894 | replisome(GO:0030894) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.1 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0018644 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.2 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.1 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |