Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
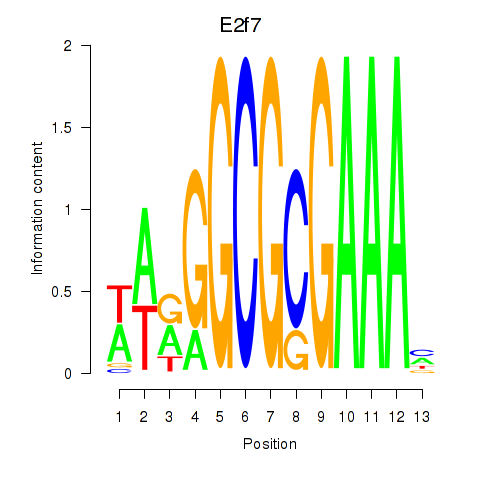
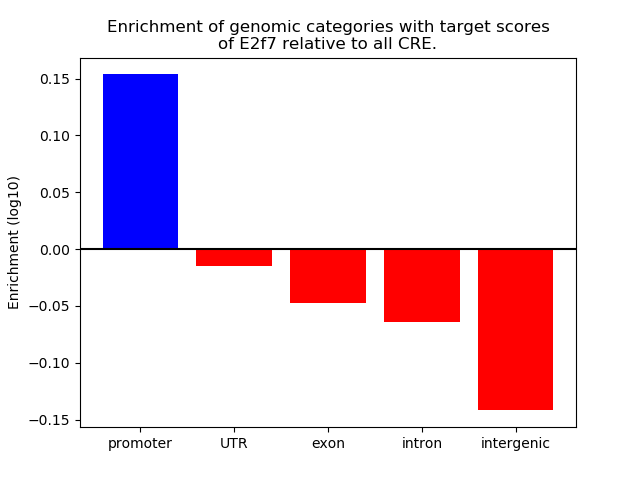
Results for E2f7
Z-value: 1.17
Transcription factors associated with E2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f7
|
ENSMUSG00000020185.10 | E2F transcription factor 7 |
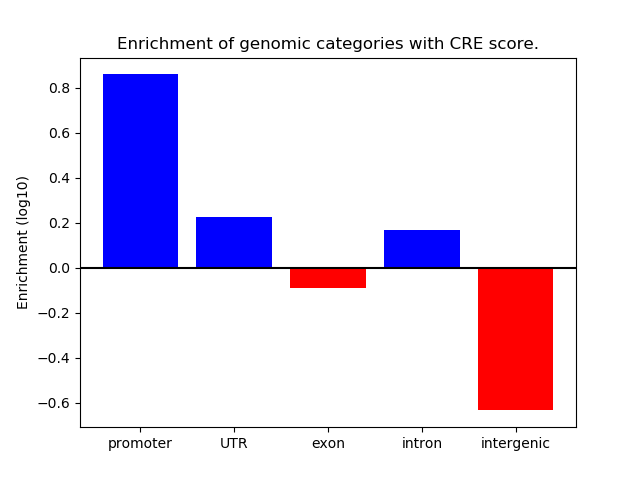
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_110706332_110706497 | E2f7 | 39025 | 0.159492 | 0.84 | 3.7e-02 | Click! |
| chr10_110717608_110717759 | E2f7 | 27756 | 0.190109 | 0.75 | 8.7e-02 | Click! |
| chr10_110707855_110708037 | E2f7 | 37493 | 0.163688 | 0.70 | 1.3e-01 | Click! |
| chr10_110727913_110728070 | E2f7 | 17448 | 0.214757 | 0.67 | 1.5e-01 | Click! |
| chr10_110717138_110717467 | E2f7 | 28137 | 0.189107 | 0.66 | 1.6e-01 | Click! |
Activity of the E2f7 motif across conditions
Conditions sorted by the z-value of the E2f7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
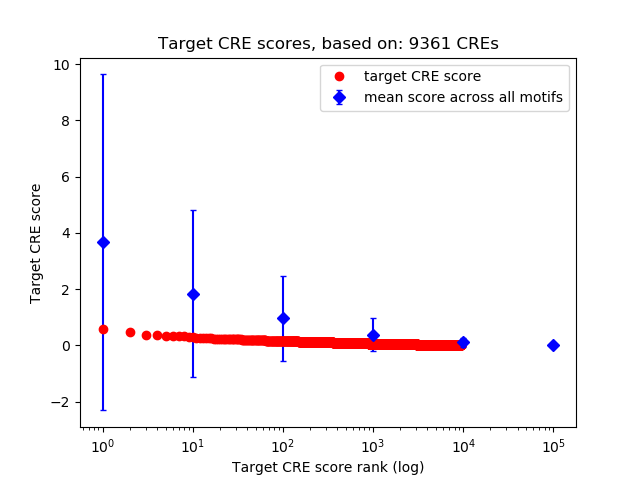
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_40356895_40357462 | 0.58 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
6087 |
0.23 |
| chr3_63976545_63977103 | 0.47 |
Gmps |
guanine monophosphate synthetase |
718 |
0.59 |
| chr14_62506079_62506243 | 0.38 |
Gm6076 |
predicted gene 6076 |
9134 |
0.13 |
| chr19_55315497_55315669 | 0.35 |
Zdhhc6 |
zinc finger, DHHC domain containing 6 |
405 |
0.66 |
| chr12_12941753_12942056 | 0.34 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
10 |
0.97 |
| chr12_104088559_104088873 | 0.33 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
8067 |
0.1 |
| chr7_99157033_99157184 | 0.32 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
12998 |
0.12 |
| chr10_3973243_3973628 | 0.32 |
Mthfd1l |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
291 |
0.88 |
| chr6_13654409_13654594 | 0.28 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
23437 |
0.21 |
| chr9_96983440_96983591 | 0.28 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
680 |
0.66 |
| chrX_113185187_113185419 | 0.28 |
Chm |
choroidermia (RAB escort protein 1) |
206 |
0.95 |
| chr9_102732216_102732808 | 0.26 |
Amotl2 |
angiomotin-like 2 |
1781 |
0.24 |
| chr2_156548213_156548364 | 0.26 |
Aar2 |
AAR2 splicing factor homolog |
664 |
0.6 |
| chr4_108425790_108426145 | 0.25 |
Gpx7 |
glutathione peroxidase 7 |
19006 |
0.1 |
| chr7_116307697_116308006 | 0.25 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
199 |
0.91 |
| chr2_12963573_12963747 | 0.24 |
Pter |
phosphotriesterase related |
14279 |
0.16 |
| chr11_90389300_90389483 | 0.24 |
Hlf |
hepatic leukemia factor |
199 |
0.96 |
| chr13_107888283_107888446 | 0.24 |
Zswim6 |
zinc finger SWIM-type containing 6 |
1700 |
0.45 |
| chr7_44815923_44816372 | 0.24 |
Nup62 |
nucleoporin 62 |
59 |
0.66 |
| chrX_12761746_12762400 | 0.24 |
Med14 |
mediator complex subunit 14 |
0 |
0.73 |
| chr3_18191923_18192090 | 0.23 |
Gm23686 |
predicted gene, 23686 |
14381 |
0.22 |
| chr1_187609763_187609931 | 0.23 |
Esrrg |
estrogen-related receptor gamma |
151 |
0.97 |
| chr1_84186357_84186508 | 0.22 |
Gm18304 |
predicted gene, 18304 |
18244 |
0.2 |
| chr15_96769798_96769949 | 0.22 |
Gm8888 |
predicted gene 8888 |
2795 |
0.32 |
| chr14_21088077_21088228 | 0.22 |
Adk |
adenosine kinase |
12000 |
0.21 |
| chr2_140774319_140774470 | 0.22 |
Flrt3 |
fibronectin leucine rich transmembrane protein 3 |
102925 |
0.08 |
| chr5_125525361_125525537 | 0.22 |
Tmem132b |
transmembrane protein 132B |
6325 |
0.17 |
| chr12_111587380_111587547 | 0.21 |
Mark3 |
MAP/microtubule affinity regulating kinase 3 |
687 |
0.55 |
| chr1_190168785_190169209 | 0.21 |
Gm28172 |
predicted gene 28172 |
327 |
0.53 |
| chr13_98263629_98263787 | 0.21 |
Ankra2 |
ankyrin repeat, family A (RFXANK-like), 2 |
466 |
0.55 |
| chr4_108143871_108144489 | 0.21 |
Scp2 |
sterol carrier protein 2, liver |
814 |
0.57 |
| chr8_13890196_13890558 | 0.21 |
Coprs |
coordinator of PRMT5, differentiation stimulator |
96 |
0.94 |
| chr7_116039656_116039813 | 0.21 |
1110004F10Rik |
RIKEN cDNA 1110004F10 gene |
22 |
0.95 |
| chr13_80892174_80892325 | 0.21 |
Arrdc3 |
arrestin domain containing 3 |
1731 |
0.32 |
| chr11_77077912_77078299 | 0.21 |
Mir423 |
microRNA 423 |
67 |
0.49 |
| chr18_44545771_44545978 | 0.21 |
Mcc |
mutated in colorectal cancers |
26358 |
0.23 |
| chr2_158865458_158865629 | 0.20 |
Dhx35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
14914 |
0.24 |
| chr11_44519752_44519982 | 0.20 |
Rnf145 |
ring finger protein 145 |
425 |
0.83 |
| chr2_11712487_11713017 | 0.20 |
Il15ra |
interleukin 15 receptor, alpha chain |
5575 |
0.16 |
| chr17_86789278_86789488 | 0.20 |
Gm50008 |
predicted gene, 50008 |
16113 |
0.17 |
| chr14_21210943_21211105 | 0.20 |
Adk |
adenosine kinase |
107101 |
0.06 |
| chrX_140493888_140494053 | 0.20 |
Prps1 |
phosphoribosyl pyrophosphate synthetase 1 |
37357 |
0.14 |
| chr11_72945131_72945282 | 0.20 |
Gm44467 |
predicted gene, 44467 |
7901 |
0.13 |
| chr17_35866032_35866679 | 0.19 |
Ppp1r18 |
protein phosphatase 1, regulatory subunit 18 |
260 |
0.6 |
| chr6_17185802_17185953 | 0.19 |
D830026I12Rik |
RIKEN cDNA D830026I12 gene |
11874 |
0.2 |
| chr8_110939430_110939581 | 0.19 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
1399 |
0.28 |
| chr11_94321337_94321604 | 0.19 |
Luc7l3 |
LUC7-like 3 (S. cerevisiae) |
425 |
0.79 |
| chr14_8326780_8326935 | 0.19 |
Fam107a |
family with sequence similarity 107, member A |
8834 |
0.19 |
| chr8_104596455_104596703 | 0.19 |
Pdp2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
5128 |
0.1 |
| chr6_121308313_121308464 | 0.19 |
Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
7606 |
0.14 |
| chr8_93277574_93277784 | 0.18 |
Ces1f |
carboxylesterase 1F |
2048 |
0.26 |
| chr6_94687247_94687564 | 0.18 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
6775 |
0.25 |
| chr2_114174645_114174811 | 0.18 |
Aqr |
aquarius |
518 |
0.79 |
| chr5_129955394_129955708 | 0.18 |
Gm43482 |
predicted gene 43482 |
8815 |
0.1 |
| chr11_97689932_97690097 | 0.18 |
Pcgf2 |
polycomb group ring finger 2 |
2393 |
0.12 |
| chr15_81871616_81871774 | 0.18 |
Phf5a |
PHD finger protein 5A |
30 |
0.9 |
| chr11_19925104_19925272 | 0.18 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
746 |
0.76 |
| chr19_11912873_11913201 | 0.18 |
Patl1 |
protein associated with topoisomerase II homolog 1 (yeast) |
594 |
0.54 |
| chr11_120231077_120231228 | 0.17 |
2900052L18Rik |
RIKEN cDNA 2900052L18 gene |
433 |
0.67 |
| chr1_21257119_21257270 | 0.17 |
Gsta3 |
glutathione S-transferase, alpha 3 |
3673 |
0.13 |
| chr9_100535718_100535869 | 0.17 |
Gm28586 |
predicted gene 28586 |
4722 |
0.16 |
| chr9_21367950_21368210 | 0.17 |
Ilf3 |
interleukin enhancer binding factor 3 |
77 |
0.92 |
| chr19_44395706_44395857 | 0.17 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
10909 |
0.14 |
| chr6_83433566_83433908 | 0.17 |
Tet3 |
tet methylcytosine dioxygenase 3 |
7895 |
0.12 |
| chr7_30094964_30095448 | 0.17 |
Zfp260 |
zinc finger protein 260 |
21 |
0.94 |
| chr3_129444095_129444246 | 0.17 |
Gm5712 |
predicted gene 5712 |
10497 |
0.17 |
| chr2_129101432_129101630 | 0.17 |
Polr1b |
polymerase (RNA) I polypeptide B |
504 |
0.68 |
| chr16_97763100_97763270 | 0.17 |
Ripk4 |
receptor-interacting serine-threonine kinase 4 |
602 |
0.75 |
| chr2_38430021_38430183 | 0.17 |
Gm13589 |
predicted gene 13589 |
6100 |
0.16 |
| chr19_21481435_21481596 | 0.17 |
Gda |
guanine deaminase |
8070 |
0.26 |
| chr5_45450154_45450875 | 0.17 |
Qdpr |
quinoid dihydropteridine reductase |
278 |
0.86 |
| chr8_40995175_40995326 | 0.17 |
Mtus1 |
mitochondrial tumor suppressor 1 |
4955 |
0.19 |
| chr9_91362060_91362437 | 0.17 |
Zic4 |
zinc finger protein of the cerebellum 4 |
165 |
0.86 |
| chr11_74638791_74638959 | 0.17 |
Cluh |
clustered mitochondria (cluA/CLU1) homolog |
10620 |
0.15 |
| chr13_31808871_31809301 | 0.16 |
Foxc1 |
forkhead box C1 |
2453 |
0.26 |
| chr7_79789753_79789904 | 0.16 |
Gm44705 |
predicted gene 44705 |
797 |
0.44 |
| chr4_9825489_9825685 | 0.16 |
Gdf6 |
growth differentiation factor 6 |
18785 |
0.21 |
| chr13_3634179_3634726 | 0.16 |
Asb13 |
ankyrin repeat and SOCS box-containing 13 |
353 |
0.85 |
| chr12_51811916_51812067 | 0.16 |
Hectd1 |
HECT domain E3 ubiquitin protein ligase 1 |
17327 |
0.18 |
| chr17_12665076_12665233 | 0.16 |
Slc22a1 |
solute carrier family 22 (organic cation transporter), member 1 |
10542 |
0.17 |
| chr6_95675108_95675466 | 0.16 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
17595 |
0.29 |
| chr2_157007239_157007486 | 0.16 |
Dsn1 |
DSN1 homolog, MIS12 kinetochore complex component |
208 |
0.9 |
| chr4_40205002_40205156 | 0.16 |
Ddx58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
3464 |
0.22 |
| chrX_140599839_140600148 | 0.16 |
Tsc22d3 |
TSC22 domain family, member 3 |
666 |
0.67 |
| chr13_21364400_21364561 | 0.16 |
Zscan12 |
zinc finger and SCAN domain containing 12 |
1195 |
0.26 |
| chr9_96652901_96653065 | 0.16 |
Rasa2 |
RAS p21 protein activator 2 |
21366 |
0.15 |
| chr5_25705756_25706065 | 0.16 |
Xrcc2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
85 |
0.96 |
| chr2_130667701_130668092 | 0.16 |
Itpa |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
31 |
0.95 |
| chr9_77776976_77777127 | 0.16 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
10909 |
0.14 |
| chr11_50388012_50388306 | 0.16 |
Hnrnph1 |
heterogeneous nuclear ribonucleoprotein H1 |
3196 |
0.18 |
| chr4_135516112_135516333 | 0.16 |
Gm25317 |
predicted gene, 25317 |
3743 |
0.16 |
| chr16_38621171_38621322 | 0.15 |
Tmem39a |
transmembrane protein 39a |
33207 |
0.12 |
| chr5_32470156_32470325 | 0.15 |
Gm43313 |
predicted gene 43313 |
3045 |
0.16 |
| chr10_67051361_67051540 | 0.15 |
Reep3 |
receptor accessory protein 3 |
14128 |
0.18 |
| chr4_105206975_105207547 | 0.15 |
Plpp3 |
phospholipid phosphatase 3 |
49914 |
0.17 |
| chr12_79969197_79969348 | 0.15 |
Gm8275 |
predicted gene 8275 |
10579 |
0.18 |
| chr9_110170831_110171182 | 0.15 |
Smarcc1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
14726 |
0.15 |
| chr9_74991465_74991644 | 0.15 |
Fam214a |
family with sequence similarity 214, member A |
15443 |
0.18 |
| chr8_88362626_88362796 | 0.15 |
Brd7 |
bromodomain containing 7 |
517 |
0.8 |
| chr16_30386539_30386690 | 0.15 |
Atp13a3 |
ATPase type 13A3 |
1871 |
0.37 |
| chr5_123720048_123720199 | 0.15 |
Zcchc8 |
zinc finger, CCHC domain containing 8 |
608 |
0.62 |
| chr15_38298706_38299133 | 0.15 |
Klf10 |
Kruppel-like factor 10 |
80 |
0.96 |
| chr14_75154835_75154992 | 0.15 |
Gm15628 |
predicted gene 15628 |
18001 |
0.14 |
| chr9_77933320_77933504 | 0.15 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
7879 |
0.15 |
| chr15_77153517_77153722 | 0.15 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
63 |
0.97 |
| chr1_74034473_74034624 | 0.15 |
Tns1 |
tensin 1 |
2585 |
0.32 |
| chrX_140457137_140457466 | 0.15 |
Prps1 |
phosphoribosyl pyrophosphate synthetase 1 |
688 |
0.7 |
| chr19_44554722_44555277 | 0.15 |
Ndufb8 |
NADH:ubiquinone oxidoreductase subunit B8 |
13 |
0.96 |
| chr17_33254646_33254995 | 0.15 |
Zfp955a |
zinc finger protein 955A |
325 |
0.83 |
| chr3_21634214_21634369 | 0.15 |
Gm17935 |
predicted gene, 17935 |
14096 |
0.22 |
| chr11_32532281_32532432 | 0.15 |
Efcab9 |
EF-hand calcium binding domain 9 |
275 |
0.84 |
| chr1_21255692_21256129 | 0.15 |
Gsta3 |
glutathione S-transferase, alpha 3 |
2389 |
0.17 |
| chr11_108254530_108254689 | 0.15 |
Gm11655 |
predicted gene 11655 |
72759 |
0.09 |
| chr12_64966293_64966672 | 0.15 |
Togaram1 |
TOG array regulator of axonemal microtubules 1 |
426 |
0.67 |
| chr13_101719143_101719316 | 0.15 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
17125 |
0.2 |
| chr5_142814483_142814698 | 0.15 |
Tnrc18 |
trinucleotide repeat containing 18 |
2790 |
0.25 |
| chr8_35257088_35257239 | 0.14 |
Gm34597 |
predicted gene, 34597 |
11684 |
0.17 |
| chr11_96887053_96887204 | 0.14 |
Gm11523 |
predicted gene 11523 |
13174 |
0.08 |
| chr3_122888651_122888824 | 0.14 |
Fabp2 |
fatty acid binding protein 2, intestinal |
6335 |
0.14 |
| chr7_101301283_101301902 | 0.14 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
435 |
0.73 |
| chr17_84740428_84740579 | 0.14 |
Lrpprc |
leucine-rich PPR-motif containing |
9319 |
0.17 |
| chr14_18369590_18369741 | 0.14 |
Ube2e1 |
ubiquitin-conjugating enzyme E2E 1 |
37806 |
0.15 |
| chr19_45805094_45805257 | 0.14 |
Kcnip2 |
Kv channel-interacting protein 2 |
1601 |
0.31 |
| chr14_74947553_74947749 | 0.14 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
215 |
0.94 |
| chr9_87740016_87740167 | 0.14 |
Gm37374 |
predicted gene, 37374 |
71 |
0.97 |
| chr4_133241348_133241936 | 0.14 |
Map3k6 |
mitogen-activated protein kinase kinase kinase 6 |
824 |
0.47 |
| chr7_29211431_29211967 | 0.14 |
Catsperg1 |
cation channel sperm associated auxiliary subunit gamma 1 |
204 |
0.86 |
| chr18_84881160_84881318 | 0.14 |
Cyb5a |
cytochrome b5 type A (microsomal) |
3638 |
0.2 |
| chr13_12650352_12650636 | 0.14 |
Gpr137b-ps |
G protein-coupled receptor 137B, pseudogene |
106 |
0.94 |
| chr8_121829970_121830295 | 0.14 |
Gm24283 |
predicted gene, 24283 |
304 |
0.64 |
| chr9_86710183_86710334 | 0.14 |
Gm37484 |
predicted gene, 37484 |
1746 |
0.22 |
| chr4_98396272_98396452 | 0.14 |
Patj |
PATJ, crumbs cell polarity complex component |
470 |
0.78 |
| chr2_127070770_127071004 | 0.14 |
Blvra |
biliverdin reductase A |
194 |
0.93 |
| chr1_78510560_78510953 | 0.14 |
Mogat1 |
monoacylglycerol O-acyltransferase 1 |
235 |
0.88 |
| chr5_92525314_92525505 | 0.14 |
Scarb2 |
scavenger receptor class B, member 2 |
18576 |
0.14 |
| chr1_131620226_131620377 | 0.14 |
Gm8532 |
predicted gene 8532 |
12668 |
0.14 |
| chr4_34886892_34887287 | 0.14 |
Zfp292 |
zinc finger protein 292 |
4129 |
0.16 |
| chr12_71042601_71042779 | 0.14 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
5651 |
0.18 |
| chr17_34114308_34114459 | 0.14 |
Brd2 |
bromodomain containing 2 |
327 |
0.65 |
| chr13_23560777_23560941 | 0.14 |
H3c6 |
H3 clustered histone 6 |
1510 |
0.11 |
| chr15_8169422_8169573 | 0.14 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
391 |
0.86 |
| chr7_4739556_4740137 | 0.14 |
Kmt5c |
lysine methyltransferase 5C |
269 |
0.73 |
| chr13_54323124_54323298 | 0.14 |
Gm48622 |
predicted gene, 48622 |
4669 |
0.18 |
| chr5_22587487_22587999 | 0.14 |
Gm9057 |
predicted gene 9057 |
23786 |
0.11 |
| chr1_21242608_21242925 | 0.14 |
Gsta3 |
glutathione S-transferase, alpha 3 |
2137 |
0.2 |
| chr11_82764474_82764643 | 0.14 |
Zfp830 |
zinc finger protein 830 |
213 |
0.5 |
| chr17_27555746_27555972 | 0.14 |
Hmga1 |
high mobility group AT-hook 1 |
638 |
0.34 |
| chr11_116477086_116477269 | 0.14 |
Prpsap1 |
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
562 |
0.6 |
| chr11_87704950_87705126 | 0.14 |
1110028F11Rik |
RIKEN cDNA 1110028F11 gene |
2915 |
0.14 |
| chr2_90536611_90536782 | 0.13 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
43951 |
0.13 |
| chr5_129701203_129701458 | 0.13 |
Septin14 |
septin 14 |
2929 |
0.16 |
| chr7_49525838_49525989 | 0.13 |
Nav2 |
neuron navigator 2 |
22279 |
0.22 |
| chr4_141325202_141325608 | 0.13 |
Epha2 |
Eph receptor A2 |
8492 |
0.1 |
| chr1_39577801_39577984 | 0.13 |
Rnf149 |
ring finger protein 149 |
487 |
0.69 |
| chr8_123040127_123040722 | 0.13 |
Ankrd11 |
ankyrin repeat domain 11 |
1596 |
0.22 |
| chr4_149980056_149980645 | 0.13 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
20915 |
0.13 |
| chr15_58942719_58942985 | 0.13 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
9044 |
0.13 |
| chr2_70798248_70798536 | 0.13 |
Tlk1 |
tousled-like kinase 1 |
26836 |
0.18 |
| chr12_100629644_100629796 | 0.13 |
Gm48585 |
predicted gene, 48585 |
2927 |
0.22 |
| chrX_48623364_48623778 | 0.13 |
Slc25a14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
7 |
0.97 |
| chr14_8003568_8003795 | 0.13 |
Abhd6 |
abhydrolase domain containing 6 |
715 |
0.66 |
| chr2_11464949_11465110 | 0.13 |
Pfkfb3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
17082 |
0.12 |
| chr11_88194414_88194594 | 0.13 |
Cuedc1 |
CUE domain containing 1 |
6683 |
0.18 |
| chr6_108456531_108456780 | 0.13 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
2048 |
0.31 |
| chr3_107808036_107808187 | 0.13 |
Gm36211 |
predicted gene, 36211 |
25428 |
0.1 |
| chr8_54925357_54925725 | 0.13 |
Gpm6a |
glycoprotein m6a |
29302 |
0.13 |
| chr1_171215629_171216025 | 0.13 |
Nr1i3 |
nuclear receptor subfamily 1, group I, member 3 |
1478 |
0.19 |
| chr15_82704114_82704796 | 0.13 |
Cyp2d38-ps |
cytochrome P450, family 2, subfamily d, member 38, pseudogene |
2810 |
0.12 |
| chr2_91636079_91636258 | 0.13 |
F2 |
coagulation factor II |
227 |
0.88 |
| chr2_75608717_75608911 | 0.13 |
Gm13655 |
predicted gene 13655 |
24568 |
0.15 |
| chr10_99264296_99264649 | 0.13 |
Dusp6 |
dual specificity phosphatase 6 |
195 |
0.9 |
| chr4_58912910_58913089 | 0.13 |
Ecpas |
Ecm29 proteasome adaptor and scaffold |
250 |
0.91 |
| chr2_39140389_39140542 | 0.13 |
Gm18791 |
predicted gene, 18791 |
13499 |
0.12 |
| chr5_9101079_9101755 | 0.13 |
Tmem243 |
transmembrane protein 243, mitochondrial |
670 |
0.68 |
| chr1_67039034_67039385 | 0.13 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
337 |
0.9 |
| chr3_32511301_32511465 | 0.13 |
Zfp639 |
zinc finger protein 639 |
487 |
0.61 |
| chr12_17401853_17402148 | 0.13 |
Gm36752 |
predicted gene, 36752 |
34618 |
0.12 |
| chr15_54612739_54612908 | 0.13 |
Mal2 |
mal, T cell differentiation protein 2 |
41631 |
0.18 |
| chr9_15307654_15307814 | 0.13 |
Taf1d |
TATA-box binding protein associated factor, RNA polymerase I, D |
8 |
0.89 |
| chr3_122729656_122729846 | 0.13 |
Pde5a |
phosphodiesterase 5A, cGMP-specific |
181 |
0.95 |
| chr14_17725657_17725828 | 0.13 |
Gm48320 |
predicted gene, 48320 |
45380 |
0.18 |
| chr15_75893944_75894142 | 0.13 |
Naprt |
nicotinate phosphoribosyltransferase |
247 |
0.81 |
| chr3_89410971_89411311 | 0.13 |
Flad1 |
flavin adenine dinucleotide synthetase 1 |
640 |
0.44 |
| chr11_100505644_100505795 | 0.13 |
Acly |
ATP citrate lyase |
6621 |
0.1 |
| chr13_24281700_24281890 | 0.13 |
Carmil1 |
capping protein regulator and myosin 1 linker 1 |
1000 |
0.43 |
| chr19_37442280_37442502 | 0.13 |
Hhex |
hematopoietically expressed homeobox |
5652 |
0.13 |
| chr7_114431752_114431903 | 0.13 |
Pde3b |
phosphodiesterase 3B, cGMP-inhibited |
16546 |
0.2 |
| chr5_142421234_142421418 | 0.13 |
Foxk1 |
forkhead box K1 |
19826 |
0.19 |
| chr3_98290887_98291082 | 0.13 |
Gm43189 |
predicted gene 43189 |
3996 |
0.17 |
| chr4_103394349_103394515 | 0.13 |
Gm12718 |
predicted gene 12718 |
11911 |
0.2 |
| chr13_117024345_117024496 | 0.13 |
Parp8 |
poly (ADP-ribose) polymerase family, member 8 |
853 |
0.64 |
| chr10_20071278_20071572 | 0.13 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
28100 |
0.19 |
| chr17_83514778_83514946 | 0.13 |
Cox7a2l |
cytochrome c oxidase subunit 7A2 like |
532 |
0.84 |
| chr7_28951879_28952178 | 0.13 |
Actn4 |
actinin alpha 4 |
2269 |
0.17 |
| chr5_24597266_24597434 | 0.12 |
Smarcd3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
361 |
0.74 |
| chr15_59062348_59062499 | 0.12 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6959 |
0.24 |
| chr7_28104816_28105003 | 0.12 |
Fcgbp |
Fc fragment of IgG binding protein |
4100 |
0.13 |
| chr5_63788784_63789107 | 0.12 |
Nwd2 |
NACHT and WD repeat domain containing 2 |
11517 |
0.21 |
| chr6_144503491_144503691 | 0.12 |
Sox5os2 |
SRY (sex determining region Y)-box 5, opposite strand 2 |
22366 |
0.26 |
| chr8_23207373_23207572 | 0.12 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
873 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.2 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.3 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.0 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.0 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.0 | GO:0071035 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.0 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.0 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.5 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |