Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
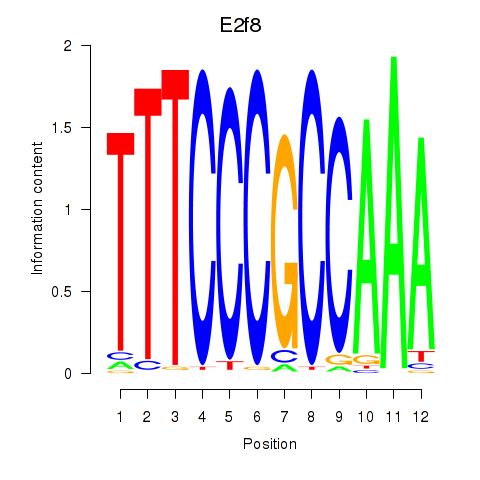
Results for E2f8
Z-value: 0.76
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSMUSG00000046179.11 | E2F transcription factor 8 |
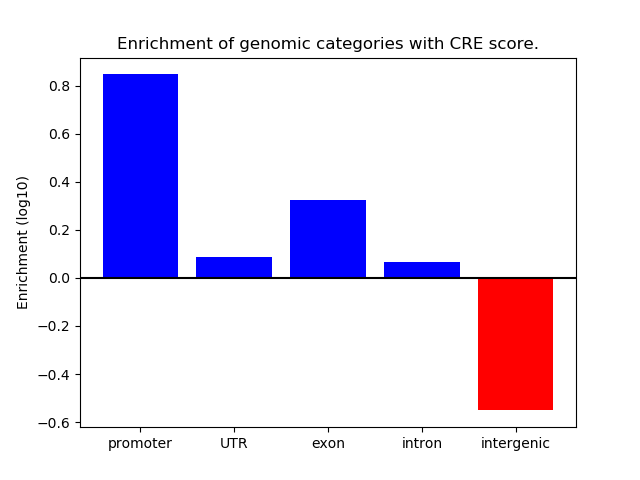
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
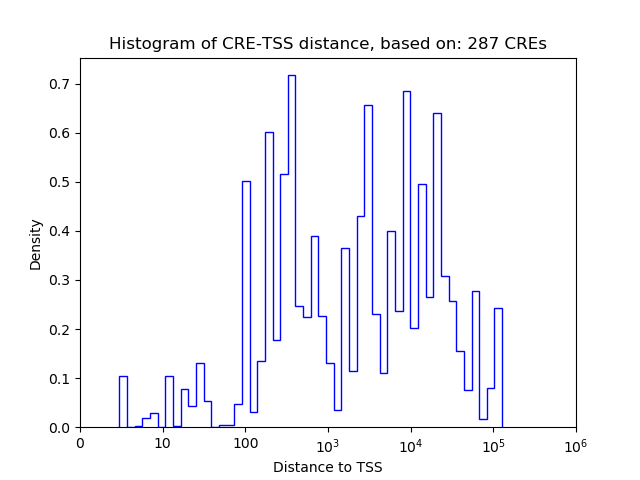
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_48881843_48881994 | E2f8 | 322 | 0.671583 | 0.71 | 1.1e-01 | Click! |
| chr7_48881042_48881203 | E2f8 | 81 | 0.634327 | -0.70 | 1.2e-01 | Click! |
| chr7_48876389_48876556 | E2f8 | 1221 | 0.312305 | 0.67 | 1.5e-01 | Click! |
| chr7_48874212_48874416 | E2f8 | 937 | 0.459593 | 0.62 | 1.9e-01 | Click! |
| chr7_48873974_48874135 | E2f8 | 1197 | 0.368804 | -0.44 | 3.8e-01 | Click! |
Activity of the E2f8 motif across conditions
Conditions sorted by the z-value of the E2f8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
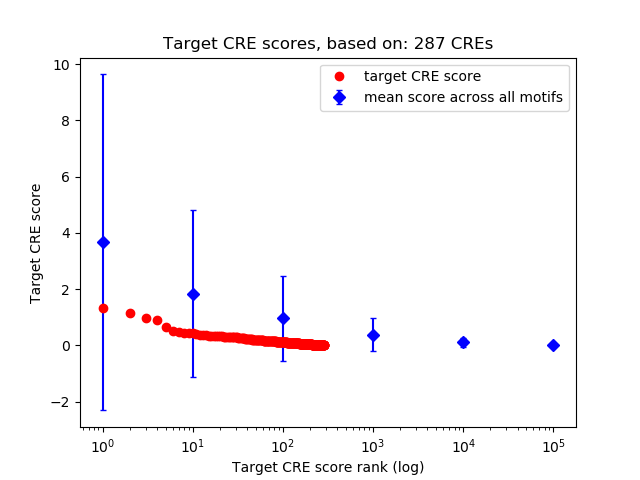
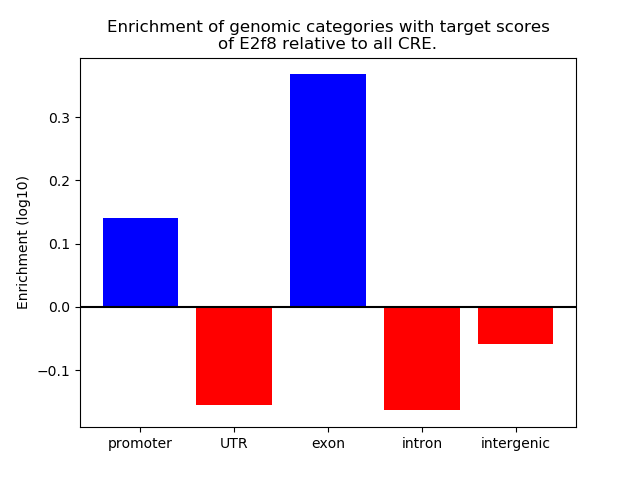
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_9825489_9825685 | 1.34 |
Gdf6 |
growth differentiation factor 6 |
18785 |
0.21 |
| chr6_149144321_149144479 | 1.13 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
2754 |
0.18 |
| chr4_137795816_137796289 | 0.96 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
178 |
0.96 |
| chr7_121981237_121981388 | 0.91 |
Cog7 |
component of oligomeric golgi complex 7 |
382 |
0.51 |
| chr2_160715771_160715932 | 0.65 |
Top1 |
topoisomerase (DNA) I |
1618 |
0.36 |
| chr2_134544863_134545049 | 0.52 |
Hao1 |
hydroxyacid oxidase 1, liver |
9351 |
0.3 |
| chr11_16817302_16817617 | 0.46 |
Egfros |
epidermal growth factor receptor, opposite strand |
13243 |
0.21 |
| chr15_98092785_98093187 | 0.44 |
Senp1 |
SUMO1/sentrin specific peptidase 1 |
99 |
0.64 |
| chr2_134795038_134795224 | 0.43 |
Plcb1 |
phospholipase C, beta 1 |
8601 |
0.19 |
| chr10_95415293_95415489 | 0.43 |
Socs2 |
suppressor of cytokine signaling 2 |
93 |
0.95 |
| chrX_140538959_140539298 | 0.42 |
Tsc22d3 |
TSC22 domain family, member 3 |
3540 |
0.25 |
| chr5_53277740_53277949 | 0.37 |
Smim20 |
small integral membrane protein 20 |
726 |
0.65 |
| chr4_3835532_3835858 | 0.37 |
Rps20 |
ribosomal protein S20 |
30 |
0.9 |
| chr1_100410526_100410677 | 0.35 |
Gm29667 |
predicted gene 29667 |
127810 |
0.05 |
| chr6_117360618_117360785 | 0.34 |
Gm4640 |
predicted gene 4640 |
62916 |
0.12 |
| chr16_34047934_34048195 | 0.34 |
Kalrn |
kalirin, RhoGEF kinase |
31570 |
0.17 |
| chr6_136906748_136907166 | 0.33 |
Erp27 |
endoplasmic reticulum protein 27 |
15183 |
0.11 |
| chr2_43522117_43522300 | 0.33 |
Gm13464 |
predicted gene 13464 |
2810 |
0.36 |
| chr9_36726566_36726754 | 0.32 |
Chek1 |
checkpoint kinase 1 |
2 |
0.96 |
| chr11_78115153_78115348 | 0.32 |
Fam222b |
family with sequence similarity 222, member B |
356 |
0.73 |
| chr5_22587487_22587999 | 0.31 |
Gm9057 |
predicted gene 9057 |
23786 |
0.11 |
| chr4_84545854_84546088 | 0.31 |
Bnc2 |
basonuclin 2 |
319 |
0.94 |
| chr11_117810052_117810409 | 0.31 |
Syngr2 |
synaptogyrin 2 |
518 |
0.39 |
| chr8_35582440_35582591 | 0.30 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
5283 |
0.21 |
| chr13_101719143_101719316 | 0.30 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
17125 |
0.2 |
| chr3_121801857_121802086 | 0.30 |
Abcd3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
12938 |
0.12 |
| chr10_87030293_87030475 | 0.29 |
Stab2 |
stabilin 2 |
22359 |
0.14 |
| chr5_124172698_124172925 | 0.29 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
12307 |
0.12 |
| chr6_54046378_54046711 | 0.28 |
Chn2 |
chimerin 2 |
6458 |
0.22 |
| chr11_29172228_29172645 | 0.28 |
Ppp4r3b |
protein phosphatase 4 regulatory subunit 3B |
454 |
0.71 |
| chr10_68147784_68148076 | 0.28 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
11304 |
0.26 |
| chr15_59072163_59072449 | 0.27 |
Mtss1 |
MTSS I-BAR domain containing 1 |
9683 |
0.23 |
| chr15_55090636_55090987 | 0.27 |
Dscc1 |
DNA replication and sister chromatid cohesion 1 |
320 |
0.85 |
| chr2_25195851_25196175 | 0.27 |
Tor4a |
torsin family 4, member A |
746 |
0.34 |
| chr2_18048778_18049088 | 0.27 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr1_175634536_175634795 | 0.27 |
Kmo |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
2426 |
0.25 |
| chr12_12941753_12942056 | 0.25 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
10 |
0.97 |
| chr3_34020715_34021194 | 0.24 |
Fxr1 |
fragile X mental retardation gene 1, autosomal homolog |
846 |
0.39 |
| chr9_94016628_94017010 | 0.23 |
Gm5369 |
predicted gene 5369 |
122643 |
0.06 |
| chr8_121953313_121953464 | 0.23 |
Gm27011 |
predicted gene, 27011 |
2743 |
0.14 |
| chr9_64281144_64281332 | 0.22 |
Tipin |
timeless interacting protein |
343 |
0.81 |
| chrX_60825498_60825671 | 0.22 |
Gm14660 |
predicted gene 14660 |
7376 |
0.21 |
| chr8_112010835_112011224 | 0.21 |
Kars |
lysyl-tRNA synthetase |
237 |
0.62 |
| chr15_82266462_82266636 | 0.21 |
Septin3 |
septin 3 |
2253 |
0.14 |
| chr11_90389300_90389483 | 0.21 |
Hlf |
hepatic leukemia factor |
199 |
0.96 |
| chr15_57693839_57694019 | 0.20 |
Zhx2 |
zinc fingers and homeoboxes 2 |
736 |
0.69 |
| chr2_44113422_44113573 | 0.20 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
48173 |
0.17 |
| chr11_80081649_80082056 | 0.20 |
Crlf3 |
cytokine receptor-like factor 3 |
861 |
0.55 |
| chr14_34188761_34188924 | 0.20 |
Gm18884 |
predicted gene, 18884 |
12533 |
0.08 |
| chr9_96823694_96824186 | 0.19 |
Gm8520 |
predicted gene 8520 |
9606 |
0.15 |
| chr17_35236084_35236374 | 0.18 |
Atp6v1g2 |
ATPase, H+ transporting, lysosomal V1 subunit G2 |
333 |
0.45 |
| chr15_99045316_99045476 | 0.18 |
Prph |
peripherin |
9778 |
0.09 |
| chr16_78481019_78481170 | 0.18 |
Gm49603 |
predicted gene, 49603 |
555 |
0.75 |
| chr4_46649810_46649996 | 0.18 |
Tbc1d2 |
TBC1 domain family, member 2 |
306 |
0.9 |
| chr18_16608064_16608271 | 0.18 |
Cdh2 |
cadherin 2 |
61902 |
0.14 |
| chr11_98979666_98979817 | 0.18 |
Gjd3 |
gap junction protein, delta 3 |
3275 |
0.14 |
| chr11_95307677_95307905 | 0.18 |
Kat7 |
K(lysine) acetyltransferase 7 |
1722 |
0.24 |
| chr19_28998112_28998308 | 0.18 |
Cdc37l1 |
cell division cycle 37-like 1 |
1107 |
0.35 |
| chr13_93996913_93997222 | 0.18 |
Gm47216 |
predicted gene, 47216 |
5299 |
0.2 |
| chr4_117131265_117131506 | 0.18 |
Plk3 |
polo like kinase 3 |
2414 |
0.1 |
| chr5_23787902_23788378 | 0.17 |
Rint1 |
RAD50 interactor 1 |
399 |
0.71 |
| chr10_47582131_47582287 | 0.17 |
Gm47571 |
predicted gene, 47571 |
57914 |
0.17 |
| chr3_136428039_136428202 | 0.16 |
1700030L20Rik |
RIKEN cDNA 1700030L20 gene |
21229 |
0.23 |
| chr15_78845338_78845568 | 0.16 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
2829 |
0.13 |
| chr4_145229848_145230015 | 0.16 |
Tnfrsf1b |
tumor necrosis factor receptor superfamily, member 1b |
16939 |
0.19 |
| chr13_112196528_112196688 | 0.16 |
Gm37427 |
predicted gene, 37427 |
22585 |
0.16 |
| chr2_84744034_84744230 | 0.16 |
Gm19426 |
predicted gene, 19426 |
477 |
0.61 |
| chr11_26386927_26387278 | 0.15 |
Fancl |
Fanconi anemia, complementation group L |
100 |
0.97 |
| chr1_67099324_67099498 | 0.15 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
23615 |
0.2 |
| chrX_150594237_150594425 | 0.15 |
Pfkfb1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
147 |
0.94 |
| chr16_70354766_70354917 | 0.15 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
40707 |
0.19 |
| chr11_79992533_79993138 | 0.15 |
Suz12 |
SUZ12 polycomb repressive complex 2 subunit |
271 |
0.91 |
| chr3_51229561_51229725 | 0.15 |
Gm38357 |
predicted gene, 38357 |
2274 |
0.22 |
| chr4_120825407_120825586 | 0.15 |
Nfyc |
nuclear transcription factor-Y gamma |
188 |
0.91 |
| chr19_40158669_40158820 | 0.15 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
28542 |
0.13 |
| chr1_192812225_192812480 | 0.15 |
Gm38360 |
predicted gene, 38360 |
8775 |
0.14 |
| chr15_75205125_75205276 | 0.15 |
Gm28117 |
predicted gene 28117 |
7533 |
0.12 |
| chr14_47242891_47243067 | 0.14 |
Wdhd1 |
WD repeat and HMG-box DNA binding protein 1 |
2291 |
0.15 |
| chr16_34824937_34825201 | 0.14 |
Mylk |
myosin, light polypeptide kinase |
40148 |
0.18 |
| chr19_6305879_6306268 | 0.14 |
Cdc42bpg |
CDC42 binding protein kinase gamma (DMPK-like) |
383 |
0.68 |
| chr7_102065637_102065828 | 0.14 |
Xntrpc |
Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
19 |
0.44 |
| chr2_165070502_165070653 | 0.14 |
Cd40 |
CD40 antigen |
7901 |
0.15 |
| chr16_38293144_38293465 | 0.14 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
1520 |
0.32 |
| chr10_10836079_10836268 | 0.13 |
4930567K20Rik |
RIKEN cDNA 4930567K20 gene |
85093 |
0.08 |
| chr4_43010254_43010416 | 0.13 |
Fancg |
Fanconi anemia, complementation group G |
20 |
0.95 |
| chr2_33370578_33371002 | 0.13 |
Ralgps1 |
Ral GEF with PH domain and SH3 binding motif 1 |
639 |
0.68 |
| chr11_55204467_55204772 | 0.13 |
Slc36a1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
243 |
0.9 |
| chr13_51073365_51073844 | 0.13 |
Spin1 |
spindlin 1 |
27276 |
0.21 |
| chr9_102721358_102721856 | 0.13 |
Amotl2 |
angiomotin-like 2 |
709 |
0.56 |
| chr10_43312960_43313230 | 0.12 |
Gm16546 |
predicted gene 16546 |
6265 |
0.19 |
| chrX_77810630_77810940 | 0.12 |
Gm14742 |
predicted gene 14742 |
7505 |
0.16 |
| chr6_145865754_145865981 | 0.12 |
Bhlhe41 |
basic helix-loop-helix family, member e41 |
309 |
0.87 |
| chr8_105881480_105881631 | 0.12 |
Edc4 |
enhancer of mRNA decapping 4 |
478 |
0.59 |
| chr8_128446539_128446690 | 0.12 |
Nrp1 |
neuropilin 1 |
87217 |
0.09 |
| chr4_108064276_108064464 | 0.11 |
Scp2 |
sterol carrier protein 2, liver |
6993 |
0.14 |
| chr18_53859942_53860093 | 0.11 |
Csnk1g3 |
casein kinase 1, gamma 3 |
2105 |
0.44 |
| chr11_31888267_31888568 | 0.11 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
15142 |
0.2 |
| chr10_19552959_19553256 | 0.11 |
Gm48563 |
predicted gene, 48563 |
30457 |
0.15 |
| chr1_51879717_51879868 | 0.11 |
Gm28323 |
predicted gene 28323 |
184 |
0.93 |
| chr17_64205783_64206178 | 0.11 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
107051 |
0.07 |
| chr1_161260611_161260770 | 0.11 |
Prdx6 |
peroxiredoxin 6 |
9471 |
0.17 |
| chr2_150778031_150778205 | 0.11 |
Pygb |
brain glycogen phosphorylase |
8617 |
0.15 |
| chr17_15423433_15423758 | 0.11 |
Gm49668 |
predicted gene, 49668 |
11369 |
0.13 |
| chr9_103349628_103349779 | 0.11 |
Topbp1 |
topoisomerase (DNA) II binding protein 1 |
3475 |
0.19 |
| chr13_21783769_21783949 | 0.11 |
H3c11 |
H3 clustered histone 11 |
489 |
0.44 |
| chr1_69108079_69108251 | 0.10 |
Erbb4 |
erb-b2 receptor tyrosine kinase 4 |
106 |
0.96 |
| chr13_89551647_89551801 | 0.10 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
11928 |
0.23 |
| chr5_20589642_20589807 | 0.10 |
Gm3544 |
predicted gene 3544 |
1128 |
0.56 |
| chr4_150740556_150740736 | 0.10 |
Gm16079 |
predicted gene 16079 |
61854 |
0.1 |
| chr16_17125381_17125599 | 0.10 |
2610318N02Rik |
RIKEN cDNA 2610318N02 gene |
323 |
0.64 |
| chr3_18624191_18624481 | 0.10 |
Gm42944 |
predicted gene 42944 |
2174 |
0.32 |
| chr16_10834941_10835111 | 0.10 |
Rmi2 |
RecQ mediated genome instability 2 |
33 |
0.95 |
| chr4_125451423_125451580 | 0.10 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
39199 |
0.16 |
| chr2_51143066_51143386 | 0.10 |
Rnd3 |
Rho family GTPase 3 |
5868 |
0.27 |
| chr1_71649974_71650188 | 0.09 |
Apol7d |
apolipoprotein L 7d |
2756 |
0.24 |
| chr8_23436686_23436866 | 0.09 |
Sfrp1 |
secreted frizzled-related protein 1 |
25274 |
0.22 |
| chr18_12861326_12861482 | 0.09 |
Osbpl1a |
oxysterol binding protein-like 1A |
199 |
0.89 |
| chr11_16820788_16820939 | 0.09 |
Egfros |
epidermal growth factor receptor, opposite strand |
9839 |
0.22 |
| chr3_96700485_96700645 | 0.09 |
Pias3 |
protein inhibitor of activated STAT 3 |
87 |
0.92 |
| chr14_99100245_99100396 | 0.09 |
Pibf1 |
progesterone immunomodulatory binding factor 1 |
362 |
0.61 |
| chr5_125173585_125173942 | 0.09 |
Ncor2 |
nuclear receptor co-repressor 2 |
5290 |
0.24 |
| chr6_83421822_83421997 | 0.09 |
5430434F05Rik |
RIKEN cDNA 5430434F05 gene |
917 |
0.44 |
| chrX_159255089_159255806 | 0.09 |
Rps6ka3 |
ribosomal protein S6 kinase polypeptide 3 |
335 |
0.92 |
| chr13_12457926_12458454 | 0.09 |
Lgals8 |
lectin, galactose binding, soluble 8 |
1667 |
0.31 |
| chr1_131276568_131276872 | 0.09 |
Ikbke |
inhibitor of kappaB kinase epsilon |
7 |
0.96 |
| chr17_32205448_32205665 | 0.09 |
Brd4 |
bromodomain containing 4 |
4548 |
0.14 |
| chr7_64497544_64497716 | 0.09 |
Apba2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
4076 |
0.21 |
| chr17_71598268_71598610 | 0.08 |
Trmt61b |
tRNA methyltransferase 61B |
186 |
0.89 |
| chr13_24316648_24316847 | 0.08 |
Gm23340 |
predicted gene, 23340 |
105 |
0.94 |
| chr16_16146290_16146715 | 0.08 |
Spidr |
scaffolding protein involved in DNA repair |
302 |
0.91 |
| chr10_17531063_17531258 | 0.08 |
Gm47770 |
predicted gene, 47770 |
4246 |
0.22 |
| chr6_116092433_116092702 | 0.08 |
Gm20404 |
predicted gene 20404 |
15083 |
0.15 |
| chr14_70351659_70351817 | 0.08 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
313 |
0.84 |
| chr17_25792594_25792983 | 0.08 |
Fam173a |
family with sequence similarity 173, member A |
10 |
0.9 |
| chr5_140783617_140783781 | 0.08 |
Amz1 |
archaelysin family metallopeptidase 1 |
34416 |
0.14 |
| chr3_95218073_95218246 | 0.08 |
Gabpb2 |
GA repeat binding protein, beta 2 |
243 |
0.72 |
| chr1_151178576_151178751 | 0.07 |
Gm47985 |
predicted gene, 47985 |
3874 |
0.14 |
| chr15_57912367_57912734 | 0.07 |
Tbc1d31 |
TBC1 domain family, member 31 |
311 |
0.9 |
| chr6_113624952_113625185 | 0.07 |
Vhl |
von Hippel-Lindau tumor suppressor |
139 |
0.89 |
| chr1_179108579_179108982 | 0.07 |
Smyd3 |
SET and MYND domain containing 3 |
119435 |
0.06 |
| chr6_15721066_15721382 | 0.07 |
Mdfic |
MyoD family inhibitor domain containing |
137 |
0.98 |
| chr18_32556472_32556888 | 0.07 |
Gypc |
glycophorin C |
3300 |
0.27 |
| chr16_33381984_33382411 | 0.07 |
Zfp148 |
zinc finger protein 148 |
579 |
0.7 |
| chr1_45795644_45795998 | 0.07 |
Wdr75 |
WD repeat domain 75 |
314 |
0.86 |
| chrX_85112549_85112727 | 0.07 |
Dmd |
dystrophin, muscular dystrophy |
64225 |
0.13 |
| chr12_111114376_111114527 | 0.07 |
Gm48632 |
predicted gene, 48632 |
23251 |
0.13 |
| chr9_57064508_57064675 | 0.07 |
Ptpn9 |
protein tyrosine phosphatase, non-receptor type 9 |
5153 |
0.14 |
| chrX_113185187_113185419 | 0.07 |
Chm |
choroidermia (RAB escort protein 1) |
206 |
0.95 |
| chr6_115717866_115718504 | 0.06 |
Gm24008 |
predicted gene, 24008 |
5647 |
0.12 |
| chr6_54051509_54051688 | 0.06 |
Chn2 |
chimerin 2 |
11512 |
0.21 |
| chr11_54860913_54861249 | 0.06 |
Lyrm7 |
LYR motif containing 7 |
165 |
0.93 |
| chr4_43000782_43000941 | 0.06 |
Vcp |
valosin containing protein |
354 |
0.76 |
| chr1_140267167_140267318 | 0.06 |
Gm37777 |
predicted gene, 37777 |
755 |
0.71 |
| chr16_7100033_7100384 | 0.06 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
30362 |
0.27 |
| chr6_37440535_37440868 | 0.06 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
1445 |
0.53 |
| chr19_55895196_55895361 | 0.06 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
232 |
0.95 |
| chr13_30338451_30338663 | 0.06 |
Agtr1a |
angiotensin II receptor, type 1a |
2113 |
0.34 |
| chr16_17619254_17619416 | 0.06 |
Smpd4 |
sphingomyelin phosphodiesterase 4 |
19 |
0.96 |
| chr3_88497845_88497996 | 0.06 |
Lmna |
lamin A |
4608 |
0.09 |
| chr2_130905756_130906591 | 0.06 |
A730017L22Rik |
RIKEN cDNA A730017L22 gene |
183 |
0.45 |
| chr17_32272780_32272931 | 0.06 |
Brd4 |
bromodomain containing 4 |
490 |
0.72 |
| chr16_92465382_92465533 | 0.06 |
Rcan1 |
regulator of calcineurin 1 |
686 |
0.61 |
| chr11_102318700_102318851 | 0.06 |
Ubtf |
upstream binding transcription factor, RNA polymerase I |
321 |
0.8 |
| chr2_17423032_17423195 | 0.05 |
Nebl |
nebulette |
32105 |
0.23 |
| chr11_79253815_79253989 | 0.05 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
568 |
0.72 |
| chr5_132546141_132546320 | 0.05 |
Gm42626 |
predicted gene 42626 |
174 |
0.91 |
| chr8_94358704_94358868 | 0.05 |
Slc12a3 |
solute carrier family 12, member 3 |
2475 |
0.16 |
| chrX_19166759_19167084 | 0.05 |
Gm14636 |
predicted gene 14636 |
311 |
0.9 |
| chr9_22003120_22003482 | 0.05 |
Prkcsh |
protein kinase C substrate 80K-H |
84 |
0.88 |
| chr5_149007925_149008200 | 0.05 |
Gm15408 |
predicted gene 15408 |
1100 |
0.3 |
| chr15_80977693_80977853 | 0.05 |
Sgsm3 |
small G protein signaling modulator 3 |
6 |
0.96 |
| chr9_32541639_32541830 | 0.05 |
Fli1 |
Friend leukemia integration 1 |
139 |
0.93 |
| chr4_95737745_95737908 | 0.05 |
Fggy |
FGGY carbohydrate kinase domain containing |
582 |
0.84 |
| chr18_74794198_74794360 | 0.05 |
Acaa2 |
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
896 |
0.53 |
| chr8_109737758_109738093 | 0.05 |
Atxn1l |
ataxin 1-like |
186 |
0.93 |
| chr12_59060973_59061324 | 0.05 |
Trappc6b |
trafficking protein particle complex 6B |
196 |
0.9 |
| chr4_137942824_137943002 | 0.04 |
Ece1 |
endothelin converting enzyme 1 |
29231 |
0.17 |
| chr10_122679401_122679658 | 0.04 |
Mir8104 |
microRNA 8104 |
214 |
0.84 |
| chr15_55557483_55557916 | 0.04 |
Mtbp |
Mdm2, transformed 3T3 cell double minute p53 binding protein |
36 |
0.51 |
| chr2_122070032_122070198 | 0.04 |
Eif3j1 |
eukaryotic translation initiation factor 3, subunit J1 |
41463 |
0.1 |
| chr11_16796275_16796460 | 0.04 |
Egfros |
epidermal growth factor receptor, opposite strand |
34335 |
0.15 |
| chr1_128102401_128102560 | 0.04 |
Zranb3 |
zinc finger, RAN-binding domain containing 3 |
17 |
0.91 |
| chr1_128359683_128359850 | 0.04 |
Mcm6 |
minichromosome maintenance complex component 6 |
102 |
0.96 |
| chr16_22263821_22263972 | 0.04 |
Tra2b |
transformer 2 beta |
2004 |
0.22 |
| chr19_37561242_37561423 | 0.04 |
Exoc6 |
exocyst complex component 6 |
10873 |
0.21 |
| chr19_46503050_46503201 | 0.04 |
Trim8 |
tripartite motif-containing 8 |
1423 |
0.35 |
| chr15_81522098_81522372 | 0.04 |
Gm5218 |
predicted gene 5218 |
22690 |
0.11 |
| chr6_122872904_122873082 | 0.04 |
Necap1 |
NECAP endocytosis associated 1 |
1481 |
0.27 |
| chr8_109991339_109991991 | 0.04 |
Tat |
tyrosine aminotransferase |
1159 |
0.36 |
| chr11_79712044_79712202 | 0.04 |
Mir193a |
microRNA 193a |
154 |
0.93 |
| chr12_80949222_80949570 | 0.04 |
Srsf5 |
serine and arginine-rich splicing factor 5 |
3020 |
0.17 |
| chr13_101349484_101349644 | 0.04 |
Gm2534 |
predicted gene 2534 |
812 |
0.69 |
| chr2_90545522_90545673 | 0.04 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
35050 |
0.16 |
| chr8_46376394_46376573 | 0.04 |
Gm45253 |
predicted gene 45253 |
9405 |
0.14 |
| chr17_4907841_4908226 | 0.03 |
4930579D07Rik |
RIKEN cDNA 4930579D07 gene |
15046 |
0.19 |
| chr1_71102457_71102615 | 0.03 |
Bard1 |
BRCA1 associated RING domain 1 |
610 |
0.8 |
| chr16_24151105_24151294 | 0.03 |
Gm32126 |
predicted gene, 32126 |
17362 |
0.19 |
| chr2_105932157_105932324 | 0.03 |
Immp1l |
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
27602 |
0.13 |
| chr2_30042566_30042728 | 0.03 |
Wdr34 |
WD repeat domain 34 |
6214 |
0.11 |
| chr13_9494911_9495081 | 0.03 |
Gm48871 |
predicted gene, 48871 |
49096 |
0.12 |
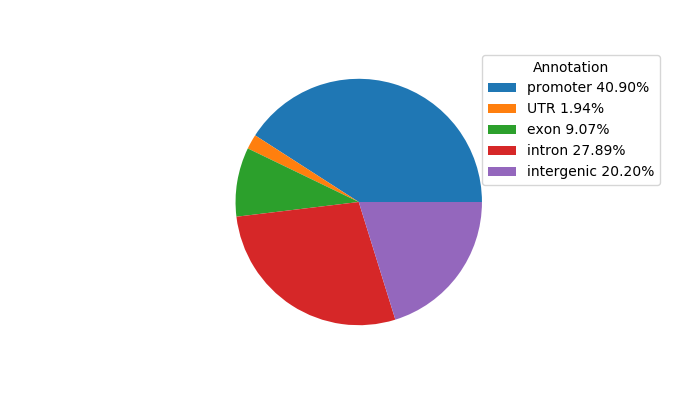
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |