Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
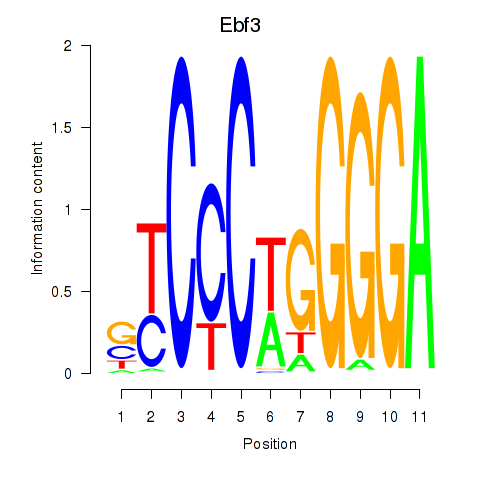
Results for Ebf3
Z-value: 1.91
Transcription factors associated with Ebf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ebf3
|
ENSMUSG00000010476.7 | early B cell factor 3 |
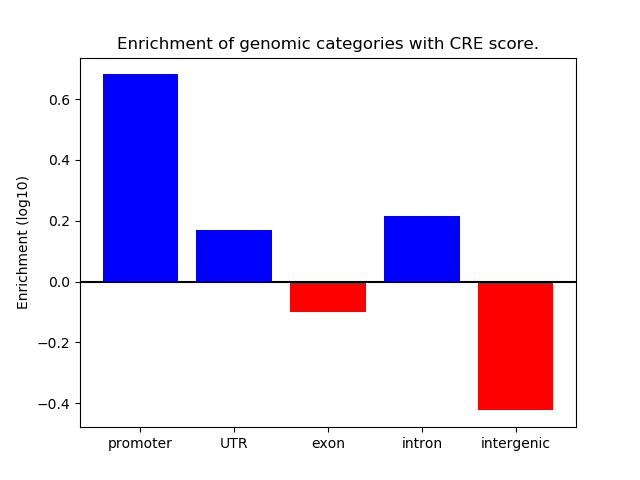
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_137319831_137319983 | Ebf3 | 5462 | 0.203989 | -0.97 | 1.0e-03 | Click! |
| chr7_137312803_137312954 | Ebf3 | 1038 | 0.541003 | -0.78 | 6.9e-02 | Click! |
| chr7_137221777_137222201 | Ebf3 | 4813 | 0.198531 | 0.73 | 9.7e-02 | Click! |
| chr7_137288505_137288677 | Ebf3 | 21053 | 0.175898 | -0.73 | 1.0e-01 | Click! |
| chr7_137223230_137223397 | Ebf3 | 3489 | 0.222007 | 0.56 | 2.5e-01 | Click! |
Activity of the Ebf3 motif across conditions
Conditions sorted by the z-value of the Ebf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
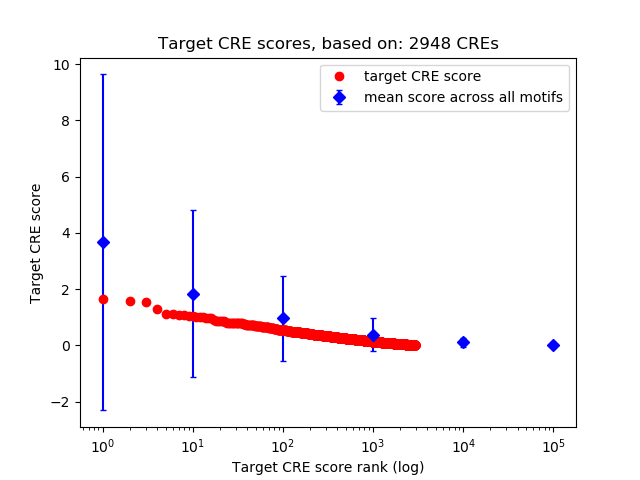
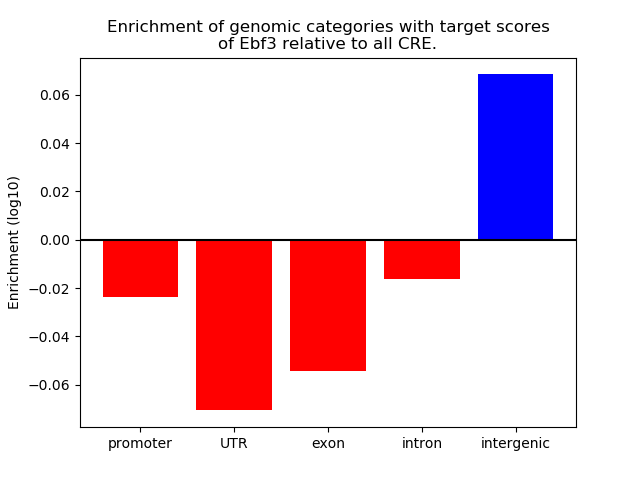
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_108330984_108331146 | 1.63 |
Apoh |
apolipoprotein H |
12289 |
0.18 |
| chr5_113152964_113153205 | 1.56 |
Gm42161 |
predicted gene, 42161 |
7756 |
0.11 |
| chr8_119416007_119416158 | 1.54 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
18042 |
0.13 |
| chr18_81199291_81199673 | 1.29 |
4930594M17Rik |
RIKEN cDNA 4930594M17 gene |
36584 |
0.17 |
| chr8_119415579_119415730 | 1.12 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
18470 |
0.13 |
| chr4_141347370_141347546 | 1.11 |
Gm13074 |
predicted gene 13074 |
356 |
0.71 |
| chr11_98767399_98767584 | 1.07 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2935 |
0.14 |
| chr10_80071687_80071929 | 1.06 |
Sbno2 |
strawberry notch 2 |
3592 |
0.11 |
| chr8_119418852_119419003 | 1.06 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
15197 |
0.14 |
| chr10_80589579_80589735 | 1.04 |
Gm29093 |
predicted gene 29093 |
133 |
0.83 |
| chr6_142459054_142459205 | 1.02 |
Gys2 |
glycogen synthase 2 |
13980 |
0.16 |
| chr17_84048784_84048940 | 1.01 |
Gm49967 |
predicted gene, 49967 |
11972 |
0.13 |
| chr19_40661223_40661381 | 0.99 |
Entpd1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
809 |
0.55 |
| chr11_75459071_75459222 | 0.99 |
Mir22hg |
Mir22 host gene (non-protein coding) |
2393 |
0.12 |
| chr2_72117636_72117826 | 0.98 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
22980 |
0.18 |
| chr19_42659380_42659560 | 0.97 |
Gm25216 |
predicted gene, 25216 |
40432 |
0.13 |
| chr1_72255173_72255368 | 0.89 |
Gm25939 |
predicted gene, 25939 |
262 |
0.86 |
| chr4_107617669_107617841 | 0.87 |
Glis1 |
GLIS family zinc finger 1 |
2755 |
0.26 |
| chr1_156996616_156996772 | 0.87 |
Gm10531 |
predicted gene 10531 |
46853 |
0.11 |
| chr5_63839973_63840346 | 0.87 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
27639 |
0.17 |
| chr4_149798966_149799139 | 0.86 |
Gm13065 |
predicted gene 13065 |
5633 |
0.1 |
| chr2_132221652_132221803 | 0.86 |
Tmem230 |
transmembrane protein 230 |
25927 |
0.13 |
| chr2_91476449_91476629 | 0.82 |
Lrp4 |
low density lipoprotein receptor-related protein 4 |
220 |
0.92 |
| chr8_119442578_119443025 | 0.81 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
3918 |
0.18 |
| chr11_101658377_101658659 | 0.81 |
Rnu2-10 |
U2 small nuclear RNA 10 |
280 |
0.81 |
| chr5_110797818_110798001 | 0.80 |
Ulk1 |
unc-51 like kinase 1 |
2075 |
0.19 |
| chr4_155245715_155246079 | 0.80 |
Faap20 |
Fanconi anemia core complex associated protein 20 |
3905 |
0.19 |
| chr10_93736289_93736487 | 0.80 |
Gm15963 |
predicted gene 15963 |
8181 |
0.16 |
| chr9_43467006_43467175 | 0.79 |
Gm28215 |
predicted gene 28215 |
2332 |
0.29 |
| chr15_72833192_72833390 | 0.79 |
Peg13 |
paternally expressed 13 |
22967 |
0.22 |
| chr3_85991904_85992061 | 0.79 |
Prss48 |
protease, serine 48 |
10509 |
0.12 |
| chr11_59885731_59885882 | 0.78 |
Gm12714 |
predicted gene 12714 |
12568 |
0.11 |
| chr19_57118911_57119062 | 0.78 |
Ablim1 |
actin-binding LIM protein 1 |
21 |
0.99 |
| chr3_95905207_95905371 | 0.78 |
Car14 |
carbonic anhydrase 14 |
598 |
0.51 |
| chr5_96222210_96222544 | 0.78 |
Mrpl1 |
mitochondrial ribosomal protein L1 |
12233 |
0.22 |
| chr14_65376633_65376799 | 0.77 |
Zfp395 |
zinc finger protein 395 |
1323 |
0.42 |
| chr12_25416673_25416856 | 0.76 |
Gm36723 |
predicted gene, 36723 |
22009 |
0.24 |
| chr7_99168829_99169197 | 0.74 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
1093 |
0.4 |
| chr9_40456362_40456513 | 0.74 |
Gramd1b |
GRAM domain containing 1B |
673 |
0.66 |
| chr5_116445735_116445902 | 0.73 |
Srrm4 |
serine/arginine repetitive matrix 4 |
1026 |
0.42 |
| chr2_50143549_50143721 | 0.73 |
Lypd6 |
LY6/PLAUR domain containing 6 |
15266 |
0.28 |
| chr5_112486568_112486719 | 0.73 |
Sez6l |
seizure related 6 homolog like |
11516 |
0.16 |
| chr6_108233581_108233762 | 0.72 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
19287 |
0.22 |
| chr14_117648617_117648937 | 0.72 |
Mir6239 |
microRNA 6239 |
305070 |
0.01 |
| chr7_112202260_112202451 | 0.72 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
23501 |
0.22 |
| chr15_102004327_102004492 | 0.71 |
Krt8 |
keratin 8 |
73 |
0.67 |
| chr1_168682011_168682173 | 0.71 |
1700063I16Rik |
RIKEN cDNA 1700063I16 gene |
6210 |
0.33 |
| chr14_20892049_20892245 | 0.71 |
Gm6128 |
predicted pseudogene 6128 |
35726 |
0.13 |
| chr5_113133182_113133354 | 0.70 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
4718 |
0.12 |
| chr13_56648096_56648259 | 0.70 |
Tgfbi |
transforming growth factor, beta induced |
16964 |
0.19 |
| chr2_174818226_174818391 | 0.70 |
Gm14617 |
predicted gene 14617 |
20326 |
0.16 |
| chr19_55229779_55229947 | 0.69 |
Gucy2g |
guanylate cyclase 2g |
11373 |
0.16 |
| chr15_82671135_82671600 | 0.69 |
Cyp2d36-ps |
cytochrome P450, family 2, subfamily d, polypeptide 36, pseudogene |
11890 |
0.08 |
| chr19_23150997_23151148 | 0.68 |
Mir1192 |
microRNA 1192 |
1641 |
0.33 |
| chr11_101952845_101953096 | 0.68 |
4930417O22Rik |
RIKEN cDNA 4930417O22 gene |
549 |
0.6 |
| chr10_128247108_128247286 | 0.68 |
Timeless |
timeless circadian clock 1 |
88 |
0.92 |
| chr14_40914867_40915027 | 0.68 |
Tspan14 |
tetraspanin 14 |
19369 |
0.18 |
| chr9_105809628_105809794 | 0.67 |
Col6a6 |
collagen, type VI, alpha 6 |
63 |
0.98 |
| chr5_36814696_36814866 | 0.66 |
Man2b2 |
mannosidase 2, alpha B2 |
1818 |
0.27 |
| chr2_30242678_30242858 | 0.66 |
Gm28035 |
predicted gene, 28035 |
2201 |
0.15 |
| chr5_125226466_125226633 | 0.66 |
Gm32585 |
predicted gene, 32585 |
46266 |
0.11 |
| chr15_66845461_66845621 | 0.66 |
Sla |
src-like adaptor |
13712 |
0.19 |
| chr1_72244368_72244519 | 0.66 |
Gm24497 |
predicted gene, 24497 |
230 |
0.88 |
| chr2_59629848_59630022 | 0.65 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
128 |
0.98 |
| chr1_172312776_172313173 | 0.65 |
Igsf8 |
immunoglobulin superfamily, member 8 |
607 |
0.57 |
| chr7_63765119_63765297 | 0.65 |
Gm44718 |
predicted gene 44718 |
666 |
0.69 |
| chr1_72226394_72226550 | 0.65 |
Gm25360 |
predicted gene, 25360 |
232 |
0.89 |
| chr5_121850762_121850913 | 0.64 |
Pheta1 |
PH domain containing endocytic trafficking adaptor 1 |
1832 |
0.19 |
| chr9_43261927_43262320 | 0.64 |
D630033O11Rik |
RIKEN cDNA D630033O11 gene |
2010 |
0.28 |
| chr4_116985890_116986058 | 0.63 |
Urod |
uroporphyrinogen decarboxylase |
5691 |
0.1 |
| chr6_136828326_136828482 | 0.62 |
Wbp11 |
WW domain binding protein 11 |
171 |
0.72 |
| chr12_111160070_111160239 | 0.62 |
Traf3 |
TNF receptor-associated factor 3 |
6216 |
0.18 |
| chr3_122120313_122120499 | 0.61 |
Abca4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
366 |
0.85 |
| chr7_27490840_27491009 | 0.61 |
Sertad1 |
SERTA domain containing 1 |
3549 |
0.11 |
| chr2_31761701_31761878 | 0.61 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
1616 |
0.33 |
| chr13_92609818_92609992 | 0.61 |
Serinc5 |
serine incorporator 5 |
1186 |
0.52 |
| chr1_191957043_191957198 | 0.60 |
Rd3 |
retinal degeneration 3 |
20250 |
0.11 |
| chr12_111514450_111514666 | 0.60 |
Gm40578 |
predicted gene, 40578 |
13718 |
0.1 |
| chr12_82086163_82086328 | 0.59 |
n-R5s62 |
nuclear encoded rRNA 5S 62 |
64223 |
0.1 |
| chr14_41101376_41101542 | 0.59 |
Mat1a |
methionine adenosyltransferase I, alpha |
3576 |
0.15 |
| chr2_124370043_124370219 | 0.59 |
4930583P06Rik |
RIKEN cDNA 4930583P06 gene |
147800 |
0.05 |
| chr16_22901028_22901197 | 0.58 |
Ahsg |
alpha-2-HS-glycoprotein |
6248 |
0.12 |
| chr19_58439431_58439649 | 0.57 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
14926 |
0.22 |
| chr5_74648783_74649196 | 0.57 |
Lnx1 |
ligand of numb-protein X 1 |
25468 |
0.16 |
| chr10_76579912_76580104 | 0.57 |
Ftcd |
formiminotransferase cyclodeaminase |
4356 |
0.13 |
| chr14_54431415_54431610 | 0.57 |
Mmp14 |
matrix metallopeptidase 14 (membrane-inserted) |
100 |
0.92 |
| chr11_101645414_101645758 | 0.57 |
Gm23971 |
predicted gene, 23971 |
344 |
0.73 |
| chr11_87662505_87662671 | 0.56 |
Rnf43 |
ring finger protein 43 |
134 |
0.94 |
| chr3_152402367_152402529 | 0.56 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
5754 |
0.12 |
| chr11_68970449_68970600 | 0.55 |
Slc25a35 |
solute carrier family 25, member 35 |
2376 |
0.12 |
| chr14_32008010_32008220 | 0.55 |
4930425P05Rik |
RIKEN cDNA 4930425P05 gene |
4887 |
0.17 |
| chr16_10366678_10366849 | 0.55 |
Gm1600 |
predicted gene 1600 |
19172 |
0.14 |
| chr11_101627513_101628017 | 0.55 |
Rdm1 |
RAD52 motif 1 |
177 |
0.69 |
| chr11_5846893_5847128 | 0.55 |
Polm |
polymerase (DNA directed), mu |
8994 |
0.11 |
| chr9_120738073_120738234 | 0.55 |
Gm47064 |
predicted gene, 47064 |
12990 |
0.13 |
| chr9_32674052_32674248 | 0.55 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
21872 |
0.15 |
| chr8_72147470_72147621 | 0.54 |
Gm26586 |
predicted gene, 26586 |
6725 |
0.08 |
| chr12_104986096_104986278 | 0.54 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
12490 |
0.12 |
| chr6_100144030_100144341 | 0.54 |
2010109P13Rik |
RIKEN cDNA 2010109P13 gene |
371 |
0.9 |
| chr5_138756463_138756623 | 0.54 |
Fam20c |
family with sequence similarity 20, member C |
1456 |
0.41 |
| chr11_101641924_101642238 | 0.54 |
Gm24950 |
predicted gene, 24950 |
333 |
0.74 |
| chr7_87938394_87938545 | 0.54 |
Gm44927 |
predicted gene 44927 |
47720 |
0.17 |
| chr1_86442760_86442964 | 0.54 |
Tex44 |
testis expressed 44 |
16533 |
0.11 |
| chr11_63827398_63827577 | 0.54 |
Gm12288 |
predicted gene 12288 |
9187 |
0.23 |
| chr9_46379532_46379689 | 0.53 |
Gm47141 |
predicted gene, 47141 |
17752 |
0.13 |
| chr2_119355713_119355881 | 0.53 |
Chac1 |
ChaC, cation transport regulator 1 |
4568 |
0.16 |
| chr5_113122736_113122907 | 0.53 |
F830115B05Rik |
RIKEN cDNA F830115B05 gene |
3357 |
0.14 |
| chr5_35737574_35737851 | 0.53 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
2275 |
0.27 |
| chr6_145679594_145679775 | 0.53 |
Tuba3b |
tubulin, alpha 3B |
63927 |
0.09 |
| chr1_74386650_74386830 | 0.52 |
Slc11a1 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
3140 |
0.13 |
| chr13_98594899_98595134 | 0.52 |
Gm4815 |
predicted gene 4815 |
18485 |
0.12 |
| chr5_33123767_33123929 | 0.52 |
Gm15936 |
predicted gene 15936 |
2954 |
0.2 |
| chr7_19866622_19866883 | 0.52 |
Gm44659 |
predicted gene 44659 |
4686 |
0.08 |
| chr1_72851647_72851798 | 0.52 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
3023 |
0.28 |
| chr12_108370797_108370948 | 0.51 |
Eml1 |
echinoderm microtubule associated protein like 1 |
85 |
0.97 |
| chr6_34727419_34727585 | 0.51 |
Npn2 |
neoplastic progression 2 |
596 |
0.66 |
| chr5_137493763_137493914 | 0.51 |
Epo |
erythropoietin |
8022 |
0.07 |
| chr5_123382074_123382225 | 0.51 |
Mlxip |
MLX interacting protein |
12649 |
0.07 |
| chr9_123529517_123529735 | 0.51 |
Sacm1l |
SAC1 suppressor of actin mutations 1-like (yeast) |
133 |
0.96 |
| chr14_20654842_20654993 | 0.51 |
Myoz1 |
myozenin 1 |
1545 |
0.21 |
| chr15_73668210_73668361 | 0.50 |
1700010B13Rik |
RIKEN cDNA 1700010B13 gene |
22411 |
0.14 |
| chr9_32657200_32657375 | 0.50 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
21039 |
0.15 |
| chr7_27607606_27607784 | 0.50 |
Akt2 |
thymoma viral proto-oncogene 2 |
53 |
0.95 |
| chr16_32124485_32124636 | 0.49 |
Gm15729 |
predicted gene 15729 |
15695 |
0.1 |
| chr2_3657810_3658181 | 0.49 |
Gm13183 |
predicted gene 13183 |
22595 |
0.15 |
| chr2_159826405_159826556 | 0.49 |
Gm11446 |
predicted gene 11446 |
32810 |
0.19 |
| chr2_27504250_27504425 | 0.48 |
Brd3 |
bromodomain containing 3 |
3325 |
0.18 |
| chr15_73492343_73492494 | 0.48 |
Gm19811 |
predicted gene, 19811 |
4879 |
0.19 |
| chr7_84145705_84145865 | 0.48 |
Abhd17c |
abhydrolase domain containing 17C |
2030 |
0.26 |
| chr3_97158666_97158826 | 0.48 |
Acp6 |
acid phosphatase 6, lysophosphatidic |
31 |
0.97 |
| chr2_166153441_166153627 | 0.48 |
Sulf2 |
sulfatase 2 |
889 |
0.57 |
| chr9_106452941_106453115 | 0.48 |
Gm28959 |
predicted gene 28959 |
110 |
0.68 |
| chr15_101260556_101260749 | 0.48 |
Nr4a1 |
nuclear receptor subfamily 4, group A, member 1 |
6194 |
0.11 |
| chr9_44129712_44129877 | 0.47 |
Mcam |
melanoma cell adhesion molecule |
4675 |
0.07 |
| chr1_84422591_84422769 | 0.47 |
Gm37959 |
predicted gene, 37959 |
27281 |
0.21 |
| chr4_128950655_128950816 | 0.47 |
Azin2 |
antizyme inhibitor 2 |
10663 |
0.15 |
| chr9_63043233_63043407 | 0.47 |
Gm24526 |
predicted gene, 24526 |
15760 |
0.17 |
| chr11_119856806_119856957 | 0.47 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
563 |
0.72 |
| chr8_126839083_126839255 | 0.47 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
64 |
0.98 |
| chr15_62712887_62713046 | 0.47 |
Gm24810 |
predicted gene, 24810 |
59962 |
0.14 |
| chr11_4031603_4031903 | 0.47 |
Sec14l4 |
SEC14-like lipid binding 4 |
30 |
0.96 |
| chr1_40117818_40118037 | 0.47 |
Gm37347 |
predicted gene, 37347 |
29416 |
0.14 |
| chr2_152817536_152817750 | 0.47 |
Bcl2l1 |
BCL2-like 1 |
10892 |
0.12 |
| chr7_45019247_45019425 | 0.46 |
Rras |
related RAS viral (r-ras) oncogene |
134 |
0.86 |
| chr9_122695637_122695788 | 0.46 |
Gm35501 |
predicted gene, 35501 |
6083 |
0.11 |
| chr5_137375760_137375924 | 0.46 |
Ephb4 |
Eph receptor B4 |
25329 |
0.08 |
| chr10_21383047_21383223 | 0.46 |
Aldh8a1 |
aldehyde dehydrogenase 8 family, member A1 |
5788 |
0.14 |
| chr19_28677852_28678003 | 0.46 |
D930032P07Rik |
RIKEN cDNA D930032P07 gene |
297 |
0.78 |
| chr12_79292996_79293163 | 0.46 |
Zfyve26 |
zinc finger, FYVE domain containing 26 |
3193 |
0.23 |
| chr7_129524479_129524630 | 0.46 |
Wdr11 |
WD repeat domain 11 |
67309 |
0.12 |
| chrX_74328709_74328905 | 0.46 |
Plxna3 |
plexin A3 |
259 |
0.78 |
| chr8_117198134_117198453 | 0.46 |
Gan |
giant axonal neuropathy |
40156 |
0.14 |
| chr11_82428957_82429108 | 0.46 |
Tmem132e |
transmembrane protein 132E |
9225 |
0.23 |
| chr16_31048299_31048452 | 0.46 |
Xxylt1 |
xyloside xylosyltransferase 1 |
13052 |
0.19 |
| chr11_89028954_89029121 | 0.46 |
Trim25 |
tripartite motif-containing 25 |
29661 |
0.09 |
| chr14_61037704_61037855 | 0.46 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
112 |
0.6 |
| chr16_23061809_23062006 | 0.46 |
Kng1 |
kininogen 1 |
3514 |
0.09 |
| chr8_119443770_119443946 | 0.45 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
2861 |
0.21 |
| chr11_117872061_117872251 | 0.45 |
Tha1 |
threonine aldolase 1 |
245 |
0.79 |
| chr9_90198554_90198732 | 0.45 |
Adamts7 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
3515 |
0.2 |
| chr6_145182126_145182277 | 0.45 |
Gm15543 |
predicted gene 15543 |
74 |
0.95 |
| chr6_108519456_108519677 | 0.45 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
834 |
0.55 |
| chr3_146617776_146618365 | 0.45 |
Dnase2b |
deoxyribonuclease II beta |
2474 |
0.2 |
| chr4_138366734_138366984 | 0.45 |
Cda |
cytidine deaminase |
1133 |
0.36 |
| chr15_97253131_97253396 | 0.45 |
Pced1b |
PC-esterase domain containing 1B |
4190 |
0.23 |
| chr15_53325064_53325247 | 0.45 |
Ext1 |
exostosin glycosyltransferase 1 |
20504 |
0.25 |
| chr2_166153665_166153816 | 0.45 |
Sulf2 |
sulfatase 2 |
683 |
0.68 |
| chr6_94640401_94640563 | 0.45 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
15043 |
0.2 |
| chr12_103658352_103658505 | 0.45 |
Serpina6 |
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
1216 |
0.31 |
| chr13_37940722_37940896 | 0.45 |
Rreb1 |
ras responsive element binding protein 1 |
6207 |
0.22 |
| chr4_105199436_105199630 | 0.45 |
Plpp3 |
phospholipid phosphatase 3 |
42186 |
0.19 |
| chr5_108674989_108675212 | 0.44 |
Slc26a1 |
solute carrier family 26 (sulfate transporter), member 1 |
315 |
0.81 |
| chr13_30660255_30660420 | 0.44 |
Dusp22 |
dual specificity phosphatase 22 |
236 |
0.94 |
| chr3_146616407_146616685 | 0.44 |
Dnase2b |
deoxyribonuclease II beta |
950 |
0.46 |
| chr4_128804978_128805273 | 0.44 |
Zfp362 |
zinc finger protein 362 |
920 |
0.55 |
| chr13_52021964_52022187 | 0.44 |
Gm37872 |
predicted gene, 37872 |
148 |
0.97 |
| chr12_36182746_36182897 | 0.44 |
Gm19046 |
predicted gene, 19046 |
10453 |
0.13 |
| chr1_46908404_46908555 | 0.43 |
Gm28527 |
predicted gene 28527 |
11600 |
0.18 |
| chr10_80917355_80917522 | 0.43 |
Lmnb2 |
lamin B2 |
413 |
0.7 |
| chr15_79734404_79734589 | 0.43 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
1455 |
0.22 |
| chr2_169115164_169115359 | 0.43 |
Gm14258 |
predicted gene 14258 |
8808 |
0.23 |
| chr13_110568602_110568764 | 0.43 |
Gm33045 |
predicted gene, 33045 |
58605 |
0.14 |
| chr6_128494689_128494877 | 0.42 |
Pzp |
PZP, alpha-2-macroglobulin like |
440 |
0.68 |
| chr4_125125048_125125199 | 0.42 |
Zc3h12a |
zinc finger CCCH type containing 12A |
2603 |
0.22 |
| chr15_96751773_96751965 | 0.42 |
Gm8888 |
predicted gene 8888 |
15209 |
0.2 |
| chr17_46031864_46032231 | 0.42 |
Vegfa |
vascular endothelial growth factor A |
232 |
0.92 |
| chr6_40634916_40635087 | 0.42 |
Mgam |
maltase-glucoamylase |
6170 |
0.17 |
| chr7_142330360_142330565 | 0.42 |
Ifitm10 |
interferon induced transmembrane protein 10 |
7560 |
0.09 |
| chr7_139366642_139366843 | 0.42 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
22367 |
0.2 |
| chr12_55473766_55473977 | 0.42 |
Nfkbia |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
17939 |
0.16 |
| chr2_32927869_32928134 | 0.42 |
Fam129b |
family with sequence similarity 129, member B |
6359 |
0.12 |
| chr2_45114016_45114230 | 0.42 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
615 |
0.66 |
| chr10_86273864_86274186 | 0.42 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
26347 |
0.19 |
| chr1_184072575_184072786 | 0.42 |
Dusp10 |
dual specificity phosphatase 10 |
38299 |
0.17 |
| chr11_98981808_98981976 | 0.42 |
Gjd3 |
gap junction protein, delta 3 |
1124 |
0.33 |
| chr9_62522367_62522518 | 0.42 |
Coro2b |
coronin, actin binding protein, 2B |
2006 |
0.36 |
| chr17_45563761_45563930 | 0.41 |
Slc35b2 |
solute carrier family 35, member B2 |
29 |
0.95 |
| chr8_119443050_119443280 | 0.41 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
3554 |
0.18 |
| chr3_96953916_96954079 | 0.41 |
Gja8 |
gap junction protein, alpha 8 |
27977 |
0.12 |
| chr16_44060984_44061135 | 0.41 |
Gramd1c |
GRAM domain containing 1C |
486 |
0.81 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 0.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.5 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.3 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.4 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.4 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.3 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.1 | 0.1 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.4 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.3 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.3 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.1 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.4 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.1 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.5 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.3 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.0 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.1 | GO:0061047 | positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.4 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.0 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.0 | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.4 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:1902218 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0046113 | purine nucleobase catabolic process(GO:0006145) xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:2000757 | negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.4 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.3 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.4 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.7 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.1 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0052744 | phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0034838 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |