Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
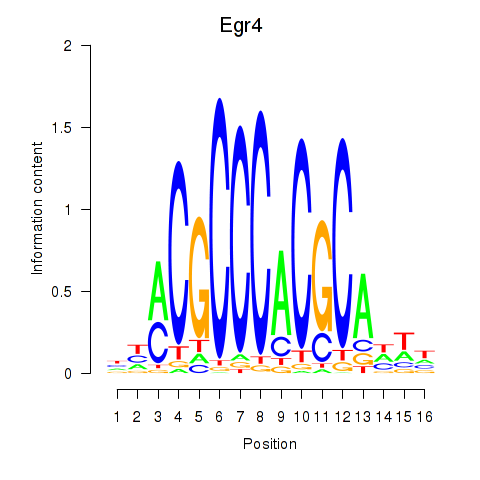
Results for Egr4
Z-value: 1.00
Transcription factors associated with Egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr4
|
ENSMUSG00000071341.3 | early growth response 4 |
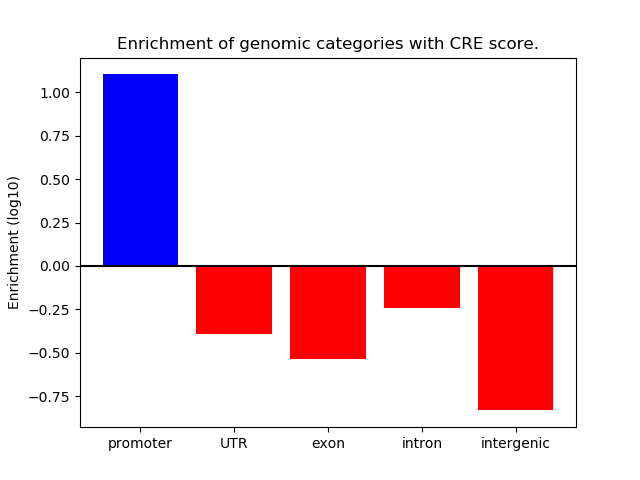
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_85510591_85510748 | Egr4 | 2920 | 0.175543 | 0.42 | 4.1e-01 | Click! |
Activity of the Egr4 motif across conditions
Conditions sorted by the z-value of the Egr4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
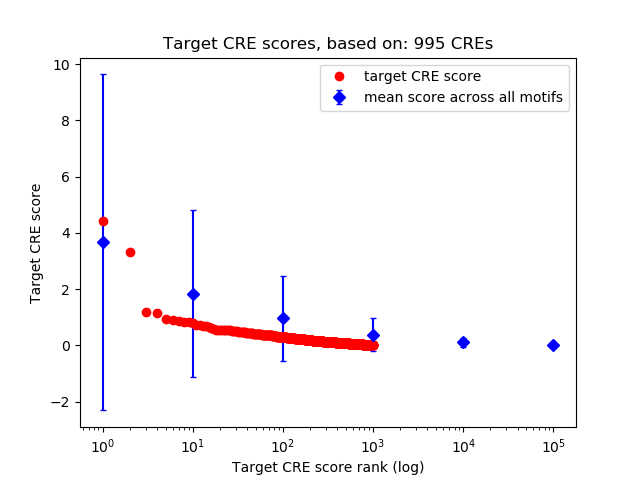
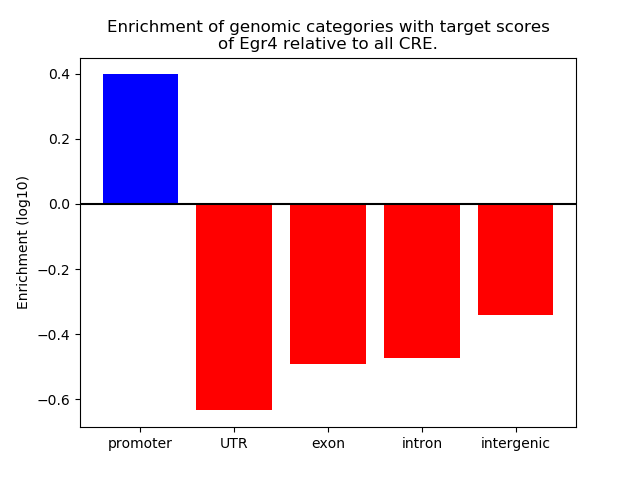
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_125569857_125570144 | 4.42 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
192 |
0.96 |
| chr9_48984872_48985421 | 3.34 |
Usp28 |
ubiquitin specific peptidase 28 |
229 |
0.92 |
| chrX_12761746_12762400 | 1.17 |
Med14 |
mediator complex subunit 14 |
0 |
0.73 |
| chr11_75173089_75173918 | 1.13 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr14_120830490_120830770 | 0.93 |
Gm9391 |
predicted gene 9391 |
3472 |
0.2 |
| chr9_89199094_89199252 | 0.90 |
Bcl2a1b |
B cell leukemia/lymphoma 2 related protein A1b |
36 |
0.71 |
| chr10_80343063_80343448 | 0.87 |
Adamtsl5 |
ADAMTS-like 5 |
1983 |
0.12 |
| chrX_9272438_9272819 | 0.84 |
Xk |
X-linked Kx blood group |
128 |
0.94 |
| chr1_182763696_182764002 | 0.82 |
Susd4 |
sushi domain containing 4 |
11 |
0.98 |
| chr9_107986066_107986266 | 0.80 |
Inka1 |
inka box actin regulator 1 |
287 |
0.67 |
| chr15_96494191_96494357 | 0.71 |
Gm41392 |
predicted gene, 41392 |
303 |
0.91 |
| chr12_76370132_76370629 | 0.71 |
Zbtb1 |
zinc finger and BTB domain containing 1 |
114 |
0.89 |
| chr19_46328692_46329074 | 0.69 |
Fbxl15 |
F-box and leucine-rich repeat protein 15 |
696 |
0.41 |
| chr8_122324060_122324293 | 0.68 |
Zfpm1 |
zinc finger protein, multitype 1 |
9522 |
0.13 |
| chrX_123103386_123103718 | 0.67 |
Cldn34c1 |
claudin 34C1 |
2 |
0.98 |
| chr3_145924039_145924997 | 0.62 |
Bcl10 |
B cell leukemia/lymphoma 10 |
161 |
0.95 |
| chr5_142938438_142938600 | 0.57 |
Fscn1 |
fascin actin-bundling protein 1 |
21824 |
0.13 |
| chr6_85431886_85432060 | 0.56 |
Smyd5 |
SET and MYND domain containing 5 |
16 |
0.96 |
| chr17_24470568_24470947 | 0.55 |
Pgp |
phosphoglycolate phosphatase |
322 |
0.51 |
| chr1_36939852_36940044 | 0.55 |
Tmem131 |
transmembrane protein 131 |
126 |
0.95 |
| chr11_97883591_97883758 | 0.55 |
Fbxo47 |
F-box protein 47 |
480 |
0.58 |
| chr10_60346558_60346709 | 0.55 |
Vsir |
V-set immunoregulatory receptor |
218 |
0.94 |
| chr3_108257311_108257615 | 0.54 |
1700010K24Rik |
RIKEN cDNA 1700010K24 gene |
478 |
0.42 |
| chr17_35667522_35667701 | 0.54 |
Vars2 |
valyl-tRNA synthetase 2, mitochondrial |
19 |
0.92 |
| chr6_128799156_128799331 | 0.53 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
10028 |
0.1 |
| chr10_79897700_79897893 | 0.53 |
Med16 |
mediator complex subunit 16 |
1889 |
0.11 |
| chr2_124609977_124610547 | 0.53 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
34 |
0.99 |
| chr15_97908200_97908485 | 0.52 |
Vdr |
vitamin D (1,25-dihydroxyvitamin D3) receptor |
32 |
0.98 |
| chr2_156008335_156008486 | 0.51 |
Ergic3 |
ERGIC and golgi 3 |
187 |
0.72 |
| chr1_153424966_153425246 | 0.51 |
Shcbp1l |
Shc SH2-domain binding protein 1-like |
56 |
0.96 |
| chr3_52267780_52268128 | 0.49 |
Foxo1 |
forkhead box O1 |
382 |
0.75 |
| chr7_25395891_25396191 | 0.49 |
Lipe |
lipase, hormone sensitive |
338 |
0.61 |
| chr9_20521562_20521965 | 0.49 |
Zfp266 |
zinc finger protein 266 |
346 |
0.81 |
| chr10_17948008_17948419 | 0.48 |
Heca |
hdc homolog, cell cycle regulator |
146 |
0.97 |
| chr2_155390028_155390312 | 0.47 |
Ncoa6 |
nuclear receptor coactivator 6 |
6656 |
0.14 |
| chr17_34958907_34959139 | 0.46 |
Hspa1b |
heat shock protein 1B |
215 |
0.75 |
| chr14_50924662_50925001 | 0.46 |
Osgep |
O-sialoglycoprotein endopeptidase |
1 |
0.62 |
| chr2_27284271_27284473 | 0.46 |
Vav2 |
vav 2 oncogene |
5493 |
0.18 |
| chr11_75170209_75170498 | 0.45 |
Hic1 |
hypermethylated in cancer 1 |
834 |
0.37 |
| chr15_79927724_79927894 | 0.45 |
Cbx7 |
chromobox 7 |
4707 |
0.14 |
| chr1_132007861_132008160 | 0.45 |
Elk4 |
ELK4, member of ETS oncogene family |
89 |
0.96 |
| chr13_58157546_58157700 | 0.44 |
Idnk |
idnK gluconokinase homolog (E. coli) |
26 |
0.51 |
| chr16_17928117_17928461 | 0.44 |
Slc25a1 |
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
70 |
0.94 |
| chr1_130715772_130716198 | 0.43 |
Pfkfb2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
171 |
0.49 |
| chr6_48086500_48086790 | 0.42 |
Zfp746 |
zinc finger protein 746 |
52 |
0.97 |
| chr11_61684235_61684526 | 0.42 |
Fam83g |
family with sequence similarity 83, member G |
39 |
0.97 |
| chr4_141060479_141060918 | 0.42 |
Crocc |
ciliary rootlet coiled-coil, rootletin |
148 |
0.92 |
| chr19_44393747_44393961 | 0.41 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12836 |
0.14 |
| chr9_55283614_55283814 | 0.40 |
Nrg4 |
neuregulin 4 |
89 |
0.97 |
| chr7_18990506_18990869 | 0.40 |
Mypop |
Myb-related transcription factor, partner of profilin |
558 |
0.49 |
| chr7_64048693_64048852 | 0.40 |
Gm45054 |
predicted gene 45054 |
7946 |
0.16 |
| chr9_88731798_88731969 | 0.40 |
Bcl2a1d |
B cell leukemia/lymphoma 2 related protein A1d |
31 |
0.59 |
| chr14_65667090_65667241 | 0.40 |
Scara5 |
scavenger receptor class A, member 5 |
728 |
0.73 |
| chr4_129618572_129618944 | 0.40 |
Iqcc |
IQ motif containing C |
318 |
0.73 |
| chr4_119138309_119138481 | 0.39 |
Gm12868 |
predicted gene 12868 |
406 |
0.71 |
| chr17_26138333_26138708 | 0.39 |
Axin1 |
axin 1 |
168 |
0.88 |
| chr3_55112219_55112827 | 0.38 |
Spg20 |
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
73 |
0.97 |
| chr6_136449342_136449720 | 0.38 |
Gm25882 |
predicted gene, 25882 |
31719 |
0.1 |
| chr17_34971805_34972034 | 0.37 |
Hspa1a |
heat shock protein 1A |
237 |
0.66 |
| chr8_75213035_75213232 | 0.37 |
Rasd2 |
RASD family, member 2 |
811 |
0.58 |
| chr15_8168911_8169078 | 0.37 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
112 |
0.97 |
| chr2_180589339_180589504 | 0.37 |
Ogfr |
opioid growth factor receptor |
41 |
0.96 |
| chr15_88832888_88833102 | 0.37 |
Gm23144 |
predicted gene, 23144 |
2305 |
0.21 |
| chr10_80577672_80578424 | 0.37 |
Klf16 |
Kruppel-like factor 16 |
727 |
0.41 |
| chr4_106463187_106463359 | 0.36 |
Pcsk9 |
proprotein convertase subtilisin/kexin type 9 |
1056 |
0.45 |
| chr7_127876515_127876679 | 0.36 |
Zfp668 |
zinc finger protein 668 |
180 |
0.49 |
| chr5_140389137_140389453 | 0.36 |
Snx8 |
sorting nexin 8 |
33 |
0.97 |
| chr6_124965028_124965351 | 0.36 |
Cops7a |
COP9 signalosome subunit 7A |
18 |
0.92 |
| chr10_77902990_77903176 | 0.36 |
Lrrc3 |
leucine rich repeat containing 3 |
547 |
0.54 |
| chr17_35186094_35186252 | 0.36 |
Lst1 |
leukocyte specific transcript 1 |
2266 |
0.07 |
| chr8_70792935_70793116 | 0.36 |
Mast3 |
microtubule associated serine/threonine kinase 3 |
588 |
0.47 |
| chr4_47352960_47353255 | 0.36 |
Tgfbr1 |
transforming growth factor, beta receptor I |
115 |
0.97 |
| chr10_4432270_4432710 | 0.35 |
Armt1 |
acidic residue methyltransferase 1 |
23 |
0.58 |
| chr4_101647910_101648061 | 0.35 |
Leprot |
leptin receptor overlapping transcript |
192 |
0.96 |
| chr4_149517672_149517903 | 0.35 |
Ctnnbip1 |
catenin beta interacting protein 1 |
449 |
0.45 |
| chr3_145790177_145790690 | 0.35 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
20046 |
0.19 |
| chr1_183345405_183345565 | 0.34 |
Mia3 |
melanoma inhibitory activity 3 |
29 |
0.97 |
| chr18_35272122_35272298 | 0.34 |
Gm50146 |
predicted gene, 50146 |
8181 |
0.17 |
| chr10_81167707_81168009 | 0.34 |
Pias4 |
protein inhibitor of activated STAT 4 |
65 |
0.92 |
| chr7_19404092_19404290 | 0.33 |
Klc3 |
kinesin light chain 3 |
87 |
0.85 |
| chr19_28834565_28834716 | 0.32 |
Slc1a1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
409 |
0.82 |
| chr18_46597468_46597778 | 0.32 |
Eif1a |
eukaryotic translation initiation factor 1A |
78 |
0.5 |
| chr13_32802244_32802687 | 0.32 |
Wrnip1 |
Werner helicase interacting protein 1 |
349 |
0.49 |
| chr11_71018794_71019067 | 0.32 |
Derl2 |
Der1-like domain family, member 2 |
282 |
0.7 |
| chr18_5819758_5819934 | 0.32 |
Zeb1 |
zinc finger E-box binding homeobox 1 |
88765 |
0.08 |
| chr1_133609970_133610131 | 0.31 |
Snrpe |
small nuclear ribonucleoprotein E |
3 |
0.97 |
| chr5_53995353_53995770 | 0.31 |
Stim2 |
stromal interaction molecule 2 |
2938 |
0.33 |
| chr5_47983016_47983183 | 0.31 |
Slit2 |
slit guidance ligand 2 |
39 |
0.97 |
| chr16_4913847_4914003 | 0.31 |
Mgrn1 |
mahogunin, ring finger 1 |
714 |
0.53 |
| chr15_76576499_76576688 | 0.31 |
Adck5 |
aarF domain containing kinase 5 |
119 |
0.88 |
| chr7_19629444_19629613 | 0.30 |
Relb |
avian reticuloendotheliosis viral (v-rel) oncogene related B |
90 |
0.93 |
| chr7_80689221_80689503 | 0.30 |
Crtc3 |
CREB regulated transcription coactivator 3 |
485 |
0.79 |
| chr4_135433737_135434028 | 0.30 |
Rcan3 |
regulator of calcineurin 3 |
29 |
0.95 |
| chr11_96075628_96076164 | 0.30 |
Atp5g1 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
226 |
0.86 |
| chr11_98005250_98005423 | 0.30 |
Cacnb1 |
calcium channel, voltage-dependent, beta 1 subunit |
7215 |
0.09 |
| chr17_31386046_31386544 | 0.29 |
Pde9a |
phosphodiesterase 9A |
1 |
0.97 |
| chr12_69680699_69681071 | 0.29 |
Sos2 |
SOS Ras/Rho guanine nucleotide exchange factor 2 |
742 |
0.62 |
| chr18_35598506_35598676 | 0.29 |
Gm50163 |
predicted gene, 50163 |
18 |
0.49 |
| chr5_98329132_98329560 | 0.29 |
Cfap299 |
cilia and flagella associated protein 299 |
5 |
0.98 |
| chr5_136244667_136245141 | 0.29 |
Sh2b2 |
SH2B adaptor protein 2 |
1 |
0.97 |
| chr1_179668115_179668496 | 0.29 |
Sccpdh |
saccharopine dehydrogenase (putative) |
45 |
0.98 |
| chr7_29211431_29211967 | 0.29 |
Catsperg1 |
cation channel sperm associated auxiliary subunit gamma 1 |
204 |
0.86 |
| chr6_137386444_137386618 | 0.29 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
24190 |
0.21 |
| chr5_99252774_99253080 | 0.29 |
Rasgef1b |
RasGEF domain family, member 1B |
0 |
0.98 |
| chr9_50659707_50659907 | 0.29 |
Dlat |
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
27 |
0.96 |
| chr12_102439744_102439900 | 0.29 |
Lgmn |
legumain |
9 |
0.98 |
| chr10_108435314_108435492 | 0.28 |
Gm36283 |
predicted gene, 36283 |
1262 |
0.46 |
| chr17_35978974_35979125 | 0.28 |
Prr3 |
proline-rich polypeptide 3 |
78 |
0.87 |
| chr7_38084831_38085137 | 0.28 |
Ccne1 |
cyclin E1 |
14385 |
0.15 |
| chr12_8674756_8674907 | 0.27 |
Pum2 |
pumilio RNA-binding family member 2 |
66 |
0.98 |
| chr14_45657978_45658304 | 0.27 |
Ddhd1 |
DDHD domain containing 1 |
1 |
0.56 |
| chr6_128760422_128760580 | 0.27 |
Gm43907 |
predicted gene, 43907 |
15857 |
0.1 |
| chr4_76360819_76360981 | 0.27 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
16657 |
0.23 |
| chr7_80370643_80370981 | 0.27 |
Man2a2 |
mannosidase 2, alpha 2 |
46 |
0.95 |
| chr6_39725291_39725442 | 0.27 |
Braf |
Braf transforming gene |
97 |
0.97 |
| chr8_94876377_94876534 | 0.27 |
Dok4 |
docking protein 4 |
125 |
0.93 |
| chr19_61226929_61227456 | 0.27 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr13_55105166_55105330 | 0.27 |
Zfp346 |
zinc finger protein 346 |
63 |
0.96 |
| chr12_5375671_5375936 | 0.27 |
2810032G03Rik |
RIKEN cDNA 2810032G03 gene |
67 |
0.57 |
| chr11_95337319_95337527 | 0.26 |
Gm11521 |
predicted gene 11521 |
15 |
0.86 |
| chr17_28523067_28523337 | 0.26 |
Gm20109 |
predicted gene, 20109 |
54 |
0.53 |
| chr13_34874226_34874546 | 0.26 |
Prpf4b |
pre-mRNA processing factor 4B |
916 |
0.34 |
| chr4_63216606_63216811 | 0.26 |
Col27a1 |
collagen, type XXVII, alpha 1 |
1273 |
0.43 |
| chr12_17691350_17691514 | 0.26 |
Hpcal1 |
hippocalcin-like 1 |
576 |
0.8 |
| chr18_67768082_67768255 | 0.26 |
Gm26910 |
predicted gene, 26910 |
6689 |
0.15 |
| chr12_110446490_110446665 | 0.26 |
Ppp2r5c |
protein phosphatase 2, regulatory subunit B', gamma |
543 |
0.76 |
| chr17_29848916_29849085 | 0.26 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
1541 |
0.36 |
| chr4_142015030_142015185 | 0.25 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
25 |
0.96 |
| chr7_80055730_80056046 | 0.25 |
Gm44951 |
predicted gene 44951 |
19183 |
0.1 |
| chr2_163750130_163750300 | 0.25 |
Ada |
adenosine deaminase |
24 |
0.98 |
| chr3_58692272_58692637 | 0.25 |
Siah2 |
siah E3 ubiquitin protein ligase 2 |
54 |
0.6 |
| chr2_169929567_169929785 | 0.25 |
AY702102 |
cDNA sequence AY702102 |
33311 |
0.2 |
| chr18_46959082_46959408 | 0.25 |
Commd10 |
COMM domain containing 10 |
365 |
0.59 |
| chr11_8663817_8664182 | 0.25 |
Tns3 |
tensin 3 |
536 |
0.87 |
| chr17_6742196_6742355 | 0.25 |
Ezr |
ezrin |
546 |
0.75 |
| chr17_56124312_56124572 | 0.24 |
Lrg1 |
leucine-rich alpha-2-glycoprotein 1 |
2441 |
0.13 |
| chr13_108158589_108158750 | 0.24 |
Ndufaf2 |
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
46 |
0.51 |
| chr3_138222614_138222780 | 0.24 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
1226 |
0.36 |
| chr15_97964312_97964525 | 0.24 |
Tmem106c |
transmembrane protein 106C |
148 |
0.95 |
| chr1_67141728_67142108 | 0.24 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
18892 |
0.22 |
| chr8_3676646_3677297 | 0.24 |
Gm45231 |
predicted gene 45231 |
17 |
0.85 |
| chr4_129600776_129601168 | 0.24 |
Tmem234 |
transmembrane protein 234 |
11 |
0.81 |
| chr4_151801848_151802061 | 0.23 |
Camta1 |
calmodulin binding transcription activator 1 |
59696 |
0.12 |
| chr4_19699207_19699375 | 0.23 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
9702 |
0.21 |
| chr18_80046692_80046991 | 0.23 |
Pard6g |
par-6 family cell polarity regulator gamma |
49 |
0.94 |
| chr6_90543682_90543913 | 0.23 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
6992 |
0.14 |
| chr9_35175976_35176147 | 0.23 |
Dcps |
decapping enzyme, scavenger |
0 |
0.96 |
| chr2_160731435_160731699 | 0.23 |
Plcg1 |
phospholipase C, gamma 1 |
132 |
0.96 |
| chr9_72132987_72133379 | 0.23 |
Gm7863 |
predicted gene 7863 |
4596 |
0.13 |
| chr7_4470400_4470562 | 0.23 |
Gm15494 |
predicted gene 15494 |
768 |
0.4 |
| chr5_99630204_99630355 | 0.23 |
4930405H06Rik |
RIKEN cDNA 4930405H06 gene |
4443 |
0.18 |
| chr17_56074581_56074743 | 0.23 |
Ubxn6 |
UBX domain protein 6 |
115 |
0.92 |
| chr8_93257060_93257211 | 0.22 |
Ces1f |
carboxylesterase 1F |
1604 |
0.3 |
| chr7_19573353_19573652 | 0.22 |
Gemin7 |
gem nuclear organelle associated protein 7 |
117 |
0.92 |
| chr19_59588162_59588313 | 0.22 |
Gm18161 |
predicted gene, 18161 |
47786 |
0.13 |
| chr15_55043394_55043818 | 0.22 |
Taf2 |
TATA-box binding protein associated factor 2 |
2560 |
0.24 |
| chr10_75407266_75407457 | 0.22 |
Upb1 |
ureidopropionase, beta |
403 |
0.84 |
| chr4_53384770_53385134 | 0.22 |
Gm12496 |
predicted gene 12496 |
19078 |
0.2 |
| chr8_92355755_92356018 | 0.22 |
Crnde |
colorectal neoplasia differentially expressed (non-protein coding) |
5 |
0.97 |
| chr11_121061638_121061789 | 0.22 |
Sectm1b |
secreted and transmembrane 1B |
1590 |
0.2 |
| chr3_94363287_94363473 | 0.22 |
C2cd4d |
C2 calcium-dependent domain containing 4D |
877 |
0.31 |
| chr2_33370578_33371002 | 0.22 |
Ralgps1 |
Ral GEF with PH domain and SH3 binding motif 1 |
639 |
0.68 |
| chrX_105079895_105080067 | 0.22 |
Pbdc1 |
polysaccharide biosynthesis domain containing 1 |
206 |
0.68 |
| chr17_69383732_69384083 | 0.22 |
Zbtb14 |
zinc finger and BTB domain containing 14 |
67 |
0.79 |
| chr15_28024715_28025200 | 0.22 |
Trio |
triple functional domain (PTPRF interacting) |
3 |
0.98 |
| chr17_83350686_83350964 | 0.22 |
Eml4 |
echinoderm microtubule associated protein like 4 |
106 |
0.98 |
| chr10_85102143_85102819 | 0.21 |
Fhl4 |
four and a half LIM domains 4 |
14 |
0.65 |
| chr5_100039849_100040017 | 0.21 |
Enoph1 |
enolase-phosphatase 1 |
52 |
0.77 |
| chr13_119597768_119597952 | 0.21 |
Tmem267 |
transmembrane protein 267 |
8790 |
0.14 |
| chr1_181144140_181144500 | 0.21 |
Nvl |
nuclear VCP-like |
116 |
0.78 |
| chr18_32163467_32163952 | 0.21 |
Map3k2 |
mitogen-activated protein kinase kinase kinase 2 |
513 |
0.7 |
| chr10_128232239_128232414 | 0.21 |
Timeless |
timeless circadian clock 1 |
192 |
0.85 |
| chr6_117168297_117168684 | 0.21 |
Cxcl12 |
chemokine (C-X-C motif) ligand 12 |
45 |
0.97 |
| chrX_134058988_134059612 | 0.21 |
Cstf2 |
cleavage stimulation factor, 3' pre-RNA subunit 2 |
11 |
0.97 |
| chr6_47758577_47758728 | 0.20 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
27722 |
0.12 |
| chr12_54734831_54734991 | 0.20 |
Gm24305 |
predicted gene, 24305 |
193 |
0.83 |
| chr2_3702714_3702883 | 0.20 |
Fam107b |
family with sequence similarity 107, member B |
1215 |
0.43 |
| chr7_27361465_27361626 | 0.20 |
Gm20479 |
predicted gene 20479 |
4126 |
0.13 |
| chr7_16028493_16028658 | 0.20 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
19346 |
0.12 |
| chr19_41482156_41482948 | 0.20 |
Lcor |
ligand dependent nuclear receptor corepressor |
85 |
0.98 |
| chr9_30137436_30137587 | 0.20 |
Gm47677 |
predicted gene, 47677 |
30064 |
0.22 |
| chr3_30855609_30856108 | 0.20 |
Gpr160 |
G protein-coupled receptor 160 |
92 |
0.96 |
| chr12_54713945_54714104 | 0.20 |
Gm23804 |
predicted gene, 23804 |
194 |
0.82 |
| chr11_117341816_117342087 | 0.19 |
Septin9 |
septin 9 |
9523 |
0.19 |
| chr17_35254463_35254614 | 0.19 |
Mir8094 |
microRNA 8094 |
3249 |
0.07 |
| chr12_98628007_98628201 | 0.19 |
Spata7 |
spermatogenesis associated 7 |
53 |
0.96 |
| chr12_102308562_102308753 | 0.19 |
Rin3 |
Ras and Rab interactor 3 |
25016 |
0.19 |
| chr6_88198537_88198710 | 0.19 |
Gata2 |
GATA binding protein 2 |
289 |
0.86 |
| chr7_24399784_24400299 | 0.19 |
Smg9 |
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
120 |
0.92 |
| chr5_3927775_3928123 | 0.19 |
Akap9 |
A kinase (PRKA) anchor protein (yotiao) 9 |
105 |
0.96 |
| chr11_65787387_65787891 | 0.19 |
Map2k4 |
mitogen-activated protein kinase kinase 4 |
646 |
0.74 |
| chr9_114731036_114731195 | 0.19 |
Cmtm6 |
CKLF-like MARVEL transmembrane domain containing 6 |
1 |
0.97 |
| chr16_32673286_32673709 | 0.19 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
298 |
0.87 |
| chr6_47772904_47773093 | 0.19 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
13376 |
0.13 |
| chr6_58610800_58610960 | 0.19 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14209 |
0.2 |
| chr8_117673801_117673966 | 0.18 |
Sdr42e1 |
short chain dehydrogenase/reductase family 42E, member 1 |
194 |
0.92 |
| chr4_97776562_97776976 | 0.18 |
Nfia |
nuclear factor I/A |
839 |
0.51 |
| chr6_47664591_47664742 | 0.18 |
Gm44141 |
predicted gene, 44141 |
29369 |
0.16 |
| chr11_22376520_22376679 | 0.18 |
Rpsa-ps3 |
ribosomal protein SA, pseudogene 3 |
9693 |
0.21 |
| chr15_82987815_82987989 | 0.18 |
Tcf20 |
transcription factor 20 |
30 |
0.53 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 0.8 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.2 | 0.5 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.5 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.2 | 0.8 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 | 0.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.3 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.2 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 3.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 1.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.1 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0060375 | mast cell differentiation(GO:0060374) regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.3 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.5 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 3.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:1900271 | regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 3.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.9 | GO:0016205 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0052859 | dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.0 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |