Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
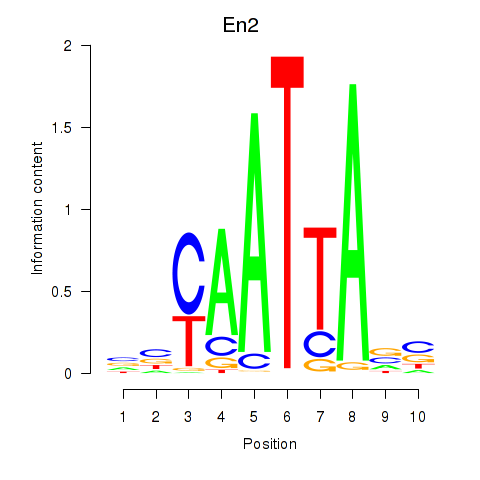
Results for En2
Z-value: 0.79
Transcription factors associated with En2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En2
|
ENSMUSG00000039095.7 | engrailed 2 |
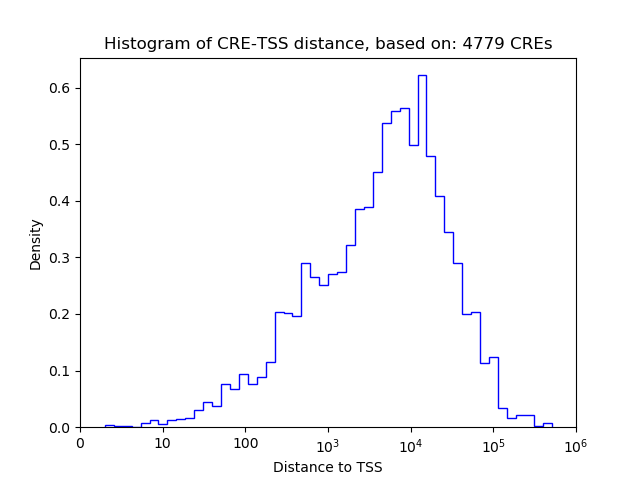
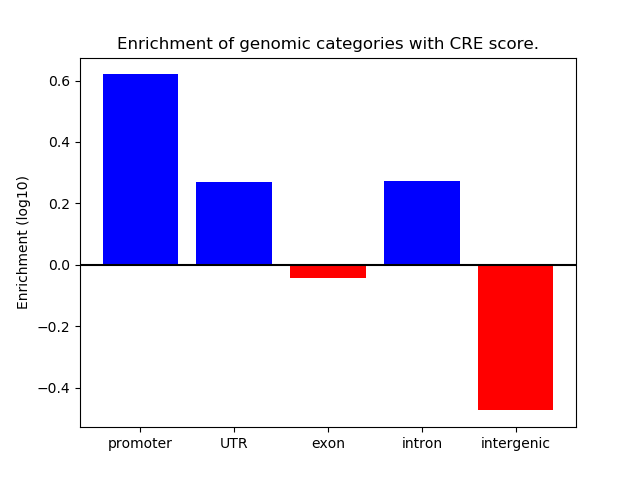
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_28123154_28123318 | En2 | 42458 | 0.112114 | 0.75 | 8.9e-02 | Click! |
Activity of the En2 motif across conditions
Conditions sorted by the z-value of the En2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
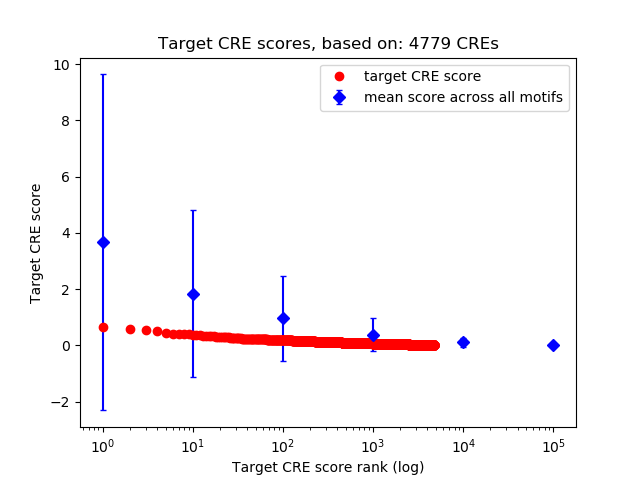
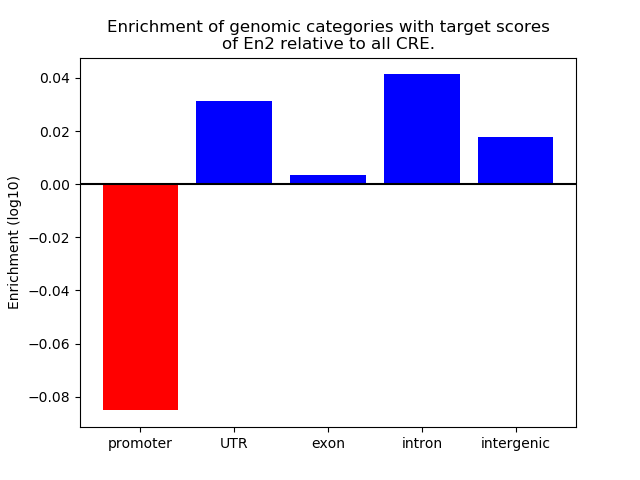
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_44400352_44400581 | 0.66 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6224 |
0.15 |
| chr16_93333790_93334136 | 0.57 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
19226 |
0.18 |
| chr17_81386211_81386362 | 0.53 |
Gm50044 |
predicted gene, 50044 |
15453 |
0.24 |
| chr19_44402814_44402965 | 0.51 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3801 |
0.18 |
| chr14_51289465_51289616 | 0.44 |
Gm49245 |
predicted gene, 49245 |
11938 |
0.1 |
| chr12_30922518_30922679 | 0.41 |
Sh3yl1 |
Sh3 domain YSC-like 1 |
10411 |
0.17 |
| chr6_58611375_58611579 | 0.40 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14806 |
0.19 |
| chr9_106788095_106788832 | 0.40 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
713 |
0.62 |
| chr9_41328613_41328764 | 0.40 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
78 |
0.97 |
| chr3_119368468_119368634 | 0.36 |
Gm23432 |
predicted gene, 23432 |
270323 |
0.02 |
| chr13_117745874_117746186 | 0.36 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr16_97905608_97905790 | 0.36 |
C2cd2 |
C2 calcium-dependent domain containing 2 |
13418 |
0.17 |
| chr8_108703572_108703757 | 0.34 |
Zfhx3 |
zinc finger homeobox 3 |
564 |
0.82 |
| chr19_44401346_44401497 | 0.34 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr10_51233946_51234117 | 0.34 |
Gm5040 |
predicted gene 5040 |
4798 |
0.28 |
| chr6_119404440_119404707 | 0.33 |
Adipor2 |
adiponectin receptor 2 |
12902 |
0.2 |
| chr12_73861884_73862127 | 0.32 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr2_18048778_18049088 | 0.30 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr14_21435070_21435221 | 0.30 |
Gm25864 |
predicted gene, 25864 |
15329 |
0.19 |
| chr1_151154930_151155204 | 0.30 |
C730036E19Rik |
RIKEN cDNA C730036E19 gene |
17033 |
0.11 |
| chr7_64513340_64513540 | 0.29 |
Gm45082 |
predicted gene 45082 |
1871 |
0.32 |
| chr1_74740134_74740308 | 0.29 |
Cyp27a1 |
cytochrome P450, family 27, subfamily a, polypeptide 1 |
3412 |
0.15 |
| chr9_115401508_115401659 | 0.29 |
Gm9487 |
predicted gene 9487 |
3566 |
0.16 |
| chr7_49523973_49524159 | 0.28 |
Nav2 |
neuron navigator 2 |
24126 |
0.22 |
| chr9_22430143_22430316 | 0.28 |
9530077C05Rik |
RIKEN cDNA 9530077C05 gene |
36 |
0.96 |
| chr5_125526040_125526191 | 0.27 |
Tmem132b |
transmembrane protein 132B |
5659 |
0.17 |
| chr10_67107041_67107217 | 0.26 |
Reep3 |
receptor accessory protein 3 |
10184 |
0.2 |
| chr4_71609659_71609810 | 0.26 |
Gm11231 |
predicted gene 11231 |
68279 |
0.13 |
| chr6_142345778_142346191 | 0.25 |
Pyroxd1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
306 |
0.86 |
| chr12_71296545_71296978 | 0.25 |
Dact1 |
dishevelled-binding antagonist of beta-catenin 1 |
13123 |
0.14 |
| chr9_70942553_70942713 | 0.25 |
Lipc |
lipase, hepatic |
7825 |
0.21 |
| chr11_119970402_119970571 | 0.24 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
10465 |
0.11 |
| chr7_30967090_30967447 | 0.24 |
Lsr |
lipolysis stimulated lipoprotein receptor |
6054 |
0.07 |
| chr1_92730752_92730936 | 0.24 |
Gm29483 |
predicted gene 29483 |
13642 |
0.14 |
| chr6_72155996_72156358 | 0.24 |
Gm38832 |
predicted gene, 38832 |
6672 |
0.15 |
| chr16_58629623_58629774 | 0.24 |
Ftdc2 |
ferritin domain containing 2 |
9041 |
0.14 |
| chr4_148143306_148143457 | 0.24 |
Mad2l2 |
MAD2 mitotic arrest deficient-like 2 |
1409 |
0.24 |
| chr13_35463284_35463451 | 0.24 |
Gm48703 |
predicted gene, 48703 |
67984 |
0.1 |
| chr19_3840572_3840723 | 0.24 |
Chka |
choline kinase alpha |
11126 |
0.09 |
| chr5_138080510_138080792 | 0.24 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr4_53104702_53105001 | 0.23 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
55044 |
0.11 |
| chr11_21713674_21713848 | 0.23 |
Wdpcp |
WD repeat containing planar cell polarity effector |
18632 |
0.2 |
| chr4_26470101_26470279 | 0.23 |
Gm11903 |
predicted gene 11903 |
27729 |
0.19 |
| chr8_46492911_46493695 | 0.23 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
471 |
0.78 |
| chr19_47313793_47313952 | 0.23 |
Sh3pxd2a |
SH3 and PX domains 2A |
879 |
0.58 |
| chr13_115653330_115653690 | 0.23 |
Gm47892 |
predicted gene, 47892 |
65989 |
0.13 |
| chrX_140538466_140538617 | 0.23 |
Tsc22d3 |
TSC22 domain family, member 3 |
4127 |
0.24 |
| chr18_44633864_44634028 | 0.22 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
27719 |
0.19 |
| chr10_59786230_59786381 | 0.22 |
Gm17059 |
predicted gene 17059 |
13949 |
0.14 |
| chr6_58611617_58611884 | 0.22 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
15079 |
0.19 |
| chr8_122697244_122697412 | 0.22 |
Gm10612 |
predicted gene 10612 |
532 |
0.59 |
| chr1_61639417_61639568 | 0.22 |
Pard3b |
par-3 family cell polarity regulator beta |
249 |
0.9 |
| chr2_67977965_67978321 | 0.22 |
Gm37964 |
predicted gene, 37964 |
79547 |
0.1 |
| chr3_130709873_130710026 | 0.22 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
505 |
0.69 |
| chr1_13368915_13369352 | 0.22 |
Ncoa2 |
nuclear receptor coactivator 2 |
3296 |
0.16 |
| chr14_68299957_68300145 | 0.22 |
Gm47212 |
predicted gene, 47212 |
81798 |
0.09 |
| chr8_67944016_67944202 | 0.22 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
7770 |
0.23 |
| chr5_122936823_122936974 | 0.21 |
Kdm2b |
lysine (K)-specific demethylase 2B |
11096 |
0.13 |
| chr9_102975773_102975938 | 0.21 |
Slco2a1 |
solute carrier organic anion transporter family, member 2a1 |
12857 |
0.17 |
| chr16_78574210_78574428 | 0.21 |
D16Ertd472e |
DNA segment, Chr 16, ERATO Doi 472, expressed |
2315 |
0.29 |
| chr1_132800530_132800731 | 0.21 |
Gm22609 |
predicted gene, 22609 |
13718 |
0.19 |
| chr13_50595486_50595664 | 0.21 |
Mir683-2 |
microRNA 683-2 |
5505 |
0.18 |
| chr2_103600204_103600386 | 0.21 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
33985 |
0.15 |
| chr10_23837916_23838078 | 0.21 |
Vnn3 |
vanin 3 |
13465 |
0.12 |
| chr18_60606060_60606525 | 0.21 |
Synpo |
synaptopodin |
3813 |
0.19 |
| chr2_77155937_77156088 | 0.21 |
Ccdc141 |
coiled-coil domain containing 141 |
14564 |
0.21 |
| chr19_42118053_42118204 | 0.21 |
Pi4k2a |
phosphatidylinositol 4-kinase type 2 alpha |
3450 |
0.14 |
| chr6_33823830_33824315 | 0.20 |
Exoc4 |
exocyst complex component 4 |
38028 |
0.2 |
| chr9_112027249_112027400 | 0.20 |
Mir128-2 |
microRNA 128-2 |
91387 |
0.08 |
| chr18_32560196_32560347 | 0.20 |
Gypc |
glycophorin C |
237 |
0.94 |
| chr14_28505648_28505933 | 0.20 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
110 |
0.97 |
| chr9_23436438_23436789 | 0.20 |
Bmper |
BMP-binding endothelial regulator |
62681 |
0.16 |
| chr4_144913207_144913382 | 0.20 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
5585 |
0.23 |
| chr5_33269323_33269474 | 0.20 |
Ctbp1 |
C-terminal binding protein 1 |
90 |
0.96 |
| chr5_89452029_89452180 | 0.20 |
Gc |
vitamin D binding protein |
5794 |
0.25 |
| chr5_44241551_44241719 | 0.20 |
Tapt1 |
transmembrane anterior posterior transformation 1 |
15009 |
0.11 |
| chr6_29828721_29828890 | 0.20 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
24955 |
0.16 |
| chr4_120076409_120076580 | 0.20 |
AL607142.1 |
novel protein |
11498 |
0.22 |
| chr4_115597563_115597714 | 0.19 |
Cyp4a32 |
cytochrome P450, family 4, subfamily a, polypeptide 32 |
3331 |
0.16 |
| chr16_38246128_38246487 | 0.19 |
Gsk3b |
glycogen synthase kinase 3 beta |
38269 |
0.11 |
| chr15_36578902_36579081 | 0.19 |
Gm44310 |
predicted gene, 44310 |
4553 |
0.16 |
| chr9_43808674_43808844 | 0.19 |
Nectin1 |
nectin cell adhesion molecule 1 |
12188 |
0.18 |
| chr10_21464742_21464927 | 0.19 |
Gm48386 |
predicted gene, 48386 |
19198 |
0.15 |
| chr13_37756552_37756712 | 0.19 |
Gm31600 |
predicted gene, 31600 |
5116 |
0.16 |
| chr4_49340688_49340844 | 0.19 |
Gm12453 |
predicted gene 12453 |
1482 |
0.35 |
| chr5_140794441_140794653 | 0.19 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
35610 |
0.14 |
| chr14_8302836_8303294 | 0.19 |
Fam107a |
family with sequence similarity 107, member A |
6712 |
0.2 |
| chr4_52516557_52516729 | 0.19 |
Vma21-ps |
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae), pseudogene |
19399 |
0.2 |
| chr13_90923293_90923612 | 0.19 |
Rps23 |
ribosomal protein S23 |
235 |
0.94 |
| chr5_121431395_121431558 | 0.19 |
Naa25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
7942 |
0.1 |
| chr14_60387250_60387782 | 0.19 |
Amer2 |
APC membrane recruitment 2 |
9230 |
0.22 |
| chr11_79792604_79792780 | 0.19 |
9130204K15Rik |
RIKEN cDNA 9130204K15 gene |
9805 |
0.18 |
| chr9_43823795_43823946 | 0.19 |
Nectin1 |
nectin cell adhesion molecule 1 |
2923 |
0.26 |
| chr3_35169015_35169202 | 0.19 |
1700017M07Rik |
RIKEN cDNA 1700017M07 gene |
107023 |
0.07 |
| chr10_42271611_42271917 | 0.19 |
Foxo3 |
forkhead box O3 |
4932 |
0.28 |
| chr9_48675501_48675665 | 0.19 |
Nnmt |
nicotinamide N-methyltransferase |
70430 |
0.11 |
| chr10_99600138_99600289 | 0.18 |
Gm20110 |
predicted gene, 20110 |
8958 |
0.19 |
| chr17_6966018_6966169 | 0.18 |
Rnaset2b |
ribonuclease T2B |
4541 |
0.13 |
| chr11_30792834_30792985 | 0.18 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr3_58685911_58686282 | 0.18 |
Siah2 |
siah E3 ubiquitin protein ligase 2 |
59 |
0.96 |
| chr9_60982764_60983106 | 0.18 |
Gm5122 |
predicted gene 5122 |
30546 |
0.14 |
| chr8_35495669_35495888 | 0.18 |
Eri1 |
exoribonuclease 1 |
245 |
0.93 |
| chr3_145992046_145992317 | 0.18 |
Syde2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
1490 |
0.39 |
| chr7_39487472_39487798 | 0.18 |
Gm28455 |
predicted gene 28455 |
1803 |
0.25 |
| chr8_45719244_45719451 | 0.18 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
23834 |
0.18 |
| chr7_144551116_144551314 | 0.18 |
Ppfia1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
1147 |
0.45 |
| chr9_108296266_108296431 | 0.18 |
Amt |
aminomethyltransferase |
505 |
0.57 |
| chr11_62409500_62409860 | 0.18 |
Ncor1 |
nuclear receptor co-repressor 1 |
8130 |
0.16 |
| chr15_89377019_89377232 | 0.18 |
Tymp |
thymidine phosphorylase |
86 |
0.89 |
| chr5_45473392_45473560 | 0.18 |
Clrn2 |
clarin 2 |
19725 |
0.11 |
| chr5_52975538_52975768 | 0.18 |
Gm30301 |
predicted gene, 30301 |
6384 |
0.16 |
| chr16_77467204_77467401 | 0.18 |
Gm37063 |
predicted gene, 37063 |
9632 |
0.12 |
| chr14_25916848_25916999 | 0.18 |
Tmem254a |
transmembrane protein 254a |
8865 |
0.12 |
| chr19_30093178_30093359 | 0.18 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
1307 |
0.48 |
| chr7_123850711_123850906 | 0.17 |
Gm23788 |
predicted gene, 23788 |
91401 |
0.08 |
| chr18_76242274_76242708 | 0.17 |
Smad2 |
SMAD family member 2 |
317 |
0.89 |
| chr12_79436149_79436313 | 0.17 |
Rad51b |
RAD51 paralog B |
108878 |
0.06 |
| chr16_76319982_76320335 | 0.17 |
Nrip1 |
nuclear receptor interacting protein 1 |
3500 |
0.3 |
| chr3_51255928_51256079 | 0.17 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr10_68327935_68328096 | 0.17 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
6873 |
0.22 |
| chr8_35216178_35216374 | 0.17 |
Gm34474 |
predicted gene, 34474 |
2362 |
0.23 |
| chr15_78839984_78840135 | 0.17 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
2565 |
0.14 |
| chr7_64802263_64802423 | 0.17 |
Fam189a1 |
family with sequence similarity 189, member A1 |
28691 |
0.19 |
| chr10_61666021_61666218 | 0.17 |
Ppa1 |
pyrophosphatase (inorganic) 1 |
2430 |
0.19 |
| chr9_70237659_70237810 | 0.17 |
Myo1e |
myosin IE |
30366 |
0.18 |
| chr17_70848513_70848664 | 0.17 |
Tgif1 |
TGFB-induced factor homeobox 1 |
1056 |
0.34 |
| chr14_26056611_26056762 | 0.17 |
Tmem254c |
transmembrane protein 254c |
8855 |
0.11 |
| chr3_27938338_27938489 | 0.17 |
Pld1 |
phospholipase D1 |
282 |
0.93 |
| chr10_69268049_69268226 | 0.17 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2500 |
0.29 |
| chr11_115244535_115244760 | 0.17 |
Gm25837 |
predicted gene, 25837 |
8631 |
0.11 |
| chr18_20969991_20970173 | 0.16 |
Rnf125 |
ring finger protein 125 |
8610 |
0.21 |
| chr14_70566111_70566293 | 0.16 |
Hr |
lysine demethylase and nuclear receptor corepressor |
10302 |
0.1 |
| chr17_3114053_3114243 | 0.16 |
Gm21926 |
predicted gene, 21926 |
602 |
0.43 |
| chr17_49304784_49304935 | 0.16 |
Gm17830 |
predicted gene, 17830 |
7382 |
0.19 |
| chr14_70530138_70530323 | 0.16 |
Lgi3 |
leucine-rich repeat LGI family, member 3 |
455 |
0.68 |
| chr6_57523099_57523250 | 0.16 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
7567 |
0.15 |
| chr8_109795573_109795879 | 0.16 |
Ap1g1 |
adaptor protein complex AP-1, gamma 1 subunit |
16820 |
0.14 |
| chr4_101198122_101198415 | 0.16 |
Gm24468 |
predicted gene, 24468 |
11504 |
0.14 |
| chr16_13411734_13411989 | 0.16 |
Gm15738 |
predicted gene 15738 |
7179 |
0.17 |
| chr9_46036951_46037323 | 0.16 |
Sik3 |
SIK family kinase 3 |
24137 |
0.13 |
| chr7_128057750_128057908 | 0.16 |
Gm22809 |
predicted gene, 22809 |
4031 |
0.09 |
| chr10_93490488_93490639 | 0.16 |
Hal |
histidine ammonia lyase |
1760 |
0.28 |
| chr2_31505484_31505674 | 0.16 |
Ass1 |
argininosuccinate synthetase 1 |
4853 |
0.2 |
| chr8_3277082_3277233 | 0.16 |
Insr |
insulin receptor |
2394 |
0.29 |
| chr6_108135683_108135868 | 0.16 |
Rpl36-ps12 |
ribosomal protein L36, pseudogene 12 |
4734 |
0.23 |
| chr9_43825824_43825990 | 0.16 |
Nectin1 |
nectin cell adhesion molecule 1 |
3924 |
0.23 |
| chr14_45588309_45588536 | 0.16 |
Gm15601 |
predicted gene 15601 |
682 |
0.52 |
| chr12_80181689_80181850 | 0.16 |
Actn1 |
actinin, alpha 1 |
3079 |
0.19 |
| chr6_140187703_140187872 | 0.16 |
Gm24174 |
predicted gene, 24174 |
21142 |
0.18 |
| chr9_122056710_122056883 | 0.16 |
Gm39465 |
predicted gene, 39465 |
5333 |
0.13 |
| chr3_14854527_14854724 | 0.16 |
Car3 |
carbonic anhydrase 3 |
8887 |
0.18 |
| chr8_109998140_109998715 | 0.16 |
Tat |
tyrosine aminotransferase |
7921 |
0.12 |
| chr10_42230294_42230483 | 0.16 |
Foxo3 |
forkhead box O3 |
27978 |
0.22 |
| chr19_40463615_40463981 | 0.16 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
12408 |
0.21 |
| chr14_101841763_101841965 | 0.16 |
Lmo7 |
LIM domain only 7 |
1045 |
0.64 |
| chr8_90831859_90832023 | 0.16 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
3085 |
0.18 |
| chr6_90499813_90499975 | 0.16 |
Gm44236 |
predicted gene, 44236 |
13408 |
0.1 |
| chr8_120351500_120351658 | 0.16 |
Gm22715 |
predicted gene, 22715 |
91970 |
0.06 |
| chr6_29841360_29841511 | 0.15 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
12325 |
0.18 |
| chr8_109996628_109997116 | 0.15 |
Tat |
tyrosine aminotransferase |
6366 |
0.13 |
| chr7_19684170_19684408 | 0.15 |
Apoc4 |
apolipoprotein C-IV |
2810 |
0.1 |
| chr6_6213107_6213286 | 0.15 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
3922 |
0.28 |
| chr15_59823011_59823164 | 0.15 |
Gm19510 |
predicted gene, 19510 |
28128 |
0.21 |
| chr7_112187893_112188109 | 0.15 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
28944 |
0.2 |
| chr15_102999275_102999447 | 0.15 |
Hoxc6 |
homeobox C6 |
1104 |
0.28 |
| chr11_120523107_120523310 | 0.15 |
Gm11789 |
predicted gene 11789 |
7346 |
0.07 |
| chr4_15212980_15213172 | 0.15 |
Tmem64 |
transmembrane protein 64 |
52755 |
0.13 |
| chr11_67535494_67535665 | 0.15 |
Gas7 |
growth arrest specific 7 |
11575 |
0.23 |
| chr4_11609051_11609282 | 0.15 |
Rad54b |
RAD54 homolog B (S. cerevisiae) |
209 |
0.92 |
| chr9_114763477_114763635 | 0.15 |
Cmtm7 |
CKLF-like MARVEL transmembrane domain containing 7 |
11895 |
0.15 |
| chr11_4098038_4098510 | 0.15 |
Mtfp1 |
mitochondrial fission process 1 |
2829 |
0.13 |
| chr15_76574596_76574756 | 0.15 |
Adck5 |
aarF domain containing kinase 5 |
1682 |
0.15 |
| chr11_88618769_88618942 | 0.15 |
Msi2 |
musashi RNA-binding protein 2 |
28708 |
0.2 |
| chr13_112473176_112473327 | 0.15 |
Il6st |
interleukin 6 signal transducer |
652 |
0.68 |
| chr15_3463467_3463618 | 0.15 |
Ghr |
growth hormone receptor |
8102 |
0.29 |
| chr17_10235141_10235292 | 0.15 |
Qk |
quaking |
21642 |
0.25 |
| chr3_142375387_142375618 | 0.15 |
Pdlim5 |
PDZ and LIM domain 5 |
16306 |
0.24 |
| chr3_95881097_95881422 | 0.15 |
Ciart |
circadian associated repressor of transcription |
13 |
0.94 |
| chr15_80683897_80684048 | 0.15 |
Fam83f |
family with sequence similarity 83, member F |
12125 |
0.12 |
| chr11_35750589_35750898 | 0.15 |
Pank3 |
pantothenate kinase 3 |
18741 |
0.16 |
| chr6_71909397_71909610 | 0.15 |
Polr1a |
polymerase (RNA) I polypeptide A |
434 |
0.58 |
| chr13_24981001_24981315 | 0.15 |
Mrs2 |
MRS2 magnesium transporter |
13641 |
0.14 |
| chr11_18900228_18900379 | 0.15 |
2900018N21Rik |
RIKEN cDNA 2900018N21 gene |
10463 |
0.16 |
| chr17_31065905_31066059 | 0.15 |
Gm25447 |
predicted gene, 25447 |
6378 |
0.14 |
| chr8_108895943_108896094 | 0.15 |
Gm20686 |
predicted gene 20686 |
25856 |
0.18 |
| chr3_55112219_55112827 | 0.15 |
Spg20 |
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
73 |
0.97 |
| chr7_118544510_118544696 | 0.15 |
Coq7 |
demethyl-Q 7 |
11247 |
0.14 |
| chr19_23309882_23310048 | 0.14 |
Gm24263 |
predicted gene, 24263 |
13636 |
0.18 |
| chr3_157729822_157729986 | 0.14 |
Gm33466 |
predicted gene, 33466 |
3496 |
0.3 |
| chr5_22549698_22549942 | 0.14 |
Orc5 |
origin recognition complex, subunit 5 |
302 |
0.68 |
| chr5_102728907_102729240 | 0.14 |
Arhgap24 |
Rho GTPase activating protein 24 |
4100 |
0.36 |
| chr8_116539027_116539192 | 0.14 |
Dynlrb2 |
dynein light chain roadblock-type 2 |
34094 |
0.19 |
| chr7_143068403_143068570 | 0.14 |
Tssc4 |
tumor-suppressing subchromosomal transferable fragment 4 |
763 |
0.46 |
| chr6_52610697_52610915 | 0.14 |
Gm44434 |
predicted gene, 44434 |
7561 |
0.15 |
| chr10_44697885_44698061 | 0.14 |
Gm47388 |
predicted gene, 47388 |
793 |
0.5 |
| chr8_41096427_41096618 | 0.14 |
Mtus1 |
mitochondrial tumor suppressor 1 |
13746 |
0.18 |
| chr7_126975670_126975892 | 0.14 |
Cdiptos |
CDIP transferase, opposite strand |
7 |
0.61 |
| chr19_9049073_9049224 | 0.14 |
Scgb1a1 |
secretoglobin, family 1A, member 1 (uteroglobin) |
38810 |
0.09 |
| chr9_48860359_48860556 | 0.14 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
24235 |
0.17 |
| chrX_103286700_103286914 | 0.14 |
4930519F16Rik |
RIKEN cDNA 4930519F16 gene |
1271 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.2 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0052205 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) |
| 0.0 | 0.0 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:0009155 | purine deoxyribonucleotide catabolic process(GO:0009155) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.0 | 0.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |