Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
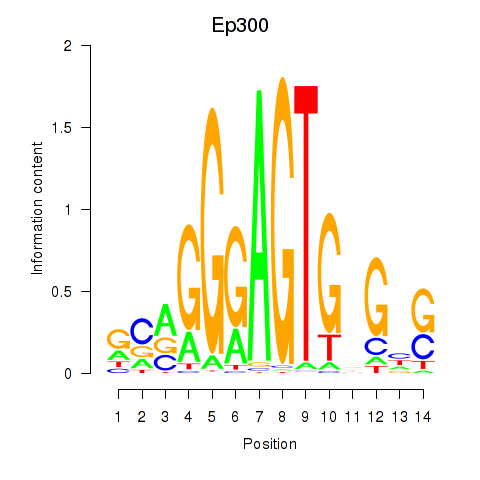
Results for Ep300
Z-value: 0.92
Transcription factors associated with Ep300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ep300
|
ENSMUSG00000055024.6 | E1A binding protein p300 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_81586343_81586511 | Ep300 | 207 | 0.574261 | 0.91 | 1.1e-02 | Click! |
| chr15_81585297_81585458 | Ep300 | 20 | 0.808049 | 0.64 | 1.7e-01 | Click! |
| chr15_81587767_81587940 | Ep300 | 1633 | 0.204495 | -0.56 | 2.5e-01 | Click! |
| chr15_81586101_81586282 | Ep300 | 29 | 0.510994 | 0.37 | 4.8e-01 | Click! |
| chr15_81586614_81586807 | Ep300 | 490 | 0.446678 | -0.35 | 4.9e-01 | Click! |
Activity of the Ep300 motif across conditions
Conditions sorted by the z-value of the Ep300 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
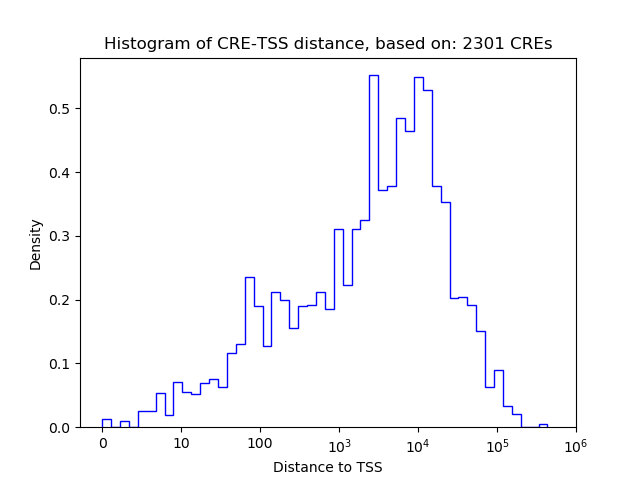
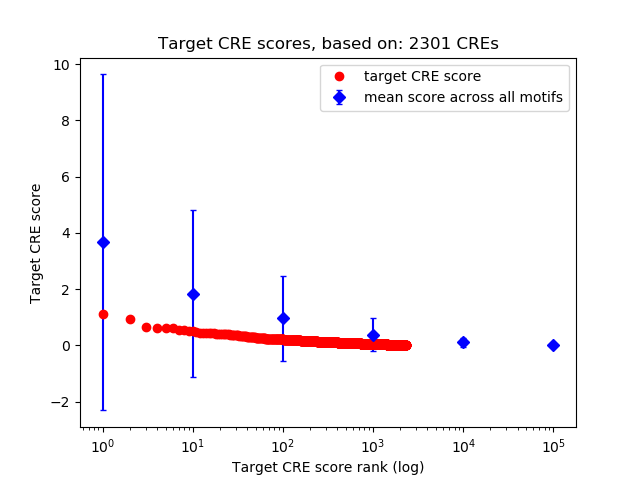
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_98148803_98148954 | 1.10 |
Omp |
olfactory marker protein |
3376 |
0.19 |
| chr19_45745506_45745808 | 0.95 |
Fgf8 |
fibroblast growth factor 8 |
2742 |
0.18 |
| chr11_4988618_4988782 | 0.66 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
1766 |
0.27 |
| chr7_127815913_127816268 | 0.63 |
Gm45204 |
predicted gene 45204 |
2583 |
0.1 |
| chr1_133908470_133908621 | 0.62 |
Optc |
opticin |
546 |
0.66 |
| chr2_118529366_118529517 | 0.61 |
Bmf |
BCL2 modifying factor |
15146 |
0.15 |
| chr11_31824504_31824681 | 0.55 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
68 |
0.97 |
| chr7_114290333_114290662 | 0.53 |
Psma1 |
proteasome subunit alpha 1 |
14379 |
0.21 |
| chr7_72498963_72499123 | 0.50 |
Gm37620 |
predicted gene, 37620 |
102946 |
0.07 |
| chr8_119444266_119444532 | 0.49 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
2320 |
0.23 |
| chr1_4479698_4479869 | 0.47 |
Sox17 |
SRY (sex determining region Y)-box 17 |
13821 |
0.14 |
| chr1_132587567_132587718 | 0.45 |
Nfasc |
neurofascin |
502 |
0.8 |
| chr4_129795439_129795602 | 0.45 |
Ptp4a2 |
protein tyrosine phosphatase 4a2 |
15699 |
0.12 |
| chr9_66243421_66243631 | 0.45 |
Dapk2 |
death-associated protein kinase 2 |
22779 |
0.2 |
| chr7_27447795_27447954 | 0.45 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
104 |
0.61 |
| chr11_6478757_6478908 | 0.44 |
Purb |
purine rich element binding protein B |
2915 |
0.11 |
| chr5_105698740_105698909 | 0.43 |
Lrrc8d |
leucine rich repeat containing 8D |
1145 |
0.57 |
| chr13_28757861_28758028 | 0.42 |
Mir6368 |
microRNA 6368 |
47071 |
0.15 |
| chr19_10266738_10266930 | 0.40 |
Dagla |
diacylglycerol lipase, alpha |
9963 |
0.13 |
| chr6_119674965_119675136 | 0.40 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
52945 |
0.15 |
| chr19_49421746_49421913 | 0.39 |
Gm50444 |
predicted gene, 50444 |
132189 |
0.06 |
| chr8_128272968_128273154 | 0.39 |
Mir21c |
microRNA 21c |
5164 |
0.3 |
| chr5_33936341_33936510 | 0.39 |
Nelfa |
negative elongation factor complex member A, Whsc2 |
12 |
0.96 |
| chr9_118944449_118944631 | 0.39 |
Ctdspl |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
14673 |
0.15 |
| chr3_9599812_9599994 | 0.39 |
Zfp704 |
zinc finger protein 704 |
10182 |
0.23 |
| chr2_69228328_69228964 | 0.38 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
8618 |
0.15 |
| chr4_155421438_155421589 | 0.38 |
5830444B04Rik |
RIKEN cDNA 5830444B04 gene |
182 |
0.71 |
| chr8_115561082_115561286 | 0.37 |
4930488N15Rik |
RIKEN cDNA 4930488N15 gene |
24208 |
0.21 |
| chr5_138979896_138980396 | 0.37 |
Pdgfa |
platelet derived growth factor, alpha |
14135 |
0.17 |
| chr15_43477084_43477240 | 0.35 |
Emc2 |
ER membrane protein complex subunit 2 |
67 |
0.58 |
| chr14_50981405_50981569 | 0.35 |
Gm8518 |
predicted gene 8518 |
2551 |
0.12 |
| chr1_164631822_164631988 | 0.35 |
Gm37754 |
predicted gene, 37754 |
148 |
0.96 |
| chr9_66802090_66802293 | 0.34 |
BC050972 |
cDNA sequence BC050972 |
368 |
0.78 |
| chr15_64163681_64163861 | 0.34 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
39965 |
0.16 |
| chr1_72827615_72827766 | 0.34 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
2368 |
0.33 |
| chr10_92235111_92235286 | 0.34 |
Gm8512 |
predicted gene 8512 |
11724 |
0.2 |
| chr12_84190226_84190393 | 0.33 |
Gm19327 |
predicted gene, 19327 |
2503 |
0.16 |
| chr3_89422230_89422381 | 0.32 |
Shc1 |
src homology 2 domain-containing transforming protein C1 |
663 |
0.44 |
| chr17_13671853_13672032 | 0.32 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
3051 |
0.19 |
| chr19_23033456_23033617 | 0.31 |
Gm50136 |
predicted gene, 50136 |
27918 |
0.18 |
| chr1_165617212_165617387 | 0.30 |
Mpzl1 |
myelin protein zero-like 1 |
3320 |
0.14 |
| chr3_97156547_97156708 | 0.30 |
Acp6 |
acid phosphatase 6, lysophosphatidic |
2150 |
0.26 |
| chr11_57983626_57984029 | 0.30 |
Gm12249 |
predicted gene 12249 |
10693 |
0.15 |
| chr11_4521480_4521785 | 0.29 |
Mtmr3 |
myotubularin related protein 3 |
6774 |
0.2 |
| chr4_133169582_133169763 | 0.29 |
Wasf2 |
WAS protein family, member 2 |
6329 |
0.15 |
| chr7_110689414_110689588 | 0.29 |
Gm21123 |
predicted gene, 21123 |
10233 |
0.15 |
| chr3_68493220_68493423 | 0.29 |
Schip1 |
schwannomin interacting protein 1 |
170 |
0.96 |
| chr16_28881932_28882083 | 0.28 |
Mb21d2 |
Mab-21 domain containing 2 |
45640 |
0.17 |
| chr19_47447791_47448053 | 0.28 |
Sh3pxd2a |
SH3 and PX domains 2A |
16325 |
0.19 |
| chr1_53931477_53931636 | 0.28 |
Gm24251 |
predicted gene, 24251 |
44615 |
0.15 |
| chr7_99164655_99164806 | 0.27 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
5376 |
0.14 |
| chr1_181860764_181860925 | 0.26 |
Gm5533 |
predicted gene 5533 |
7655 |
0.14 |
| chr5_92112862_92113049 | 0.26 |
Gm24931 |
predicted gene, 24931 |
5328 |
0.13 |
| chr6_99020013_99020438 | 0.26 |
Foxp1 |
forkhead box P1 |
7766 |
0.28 |
| chr11_62538516_62538686 | 0.26 |
Cenpv |
centromere protein V |
28 |
0.95 |
| chr15_82147771_82148080 | 0.26 |
Srebf2 |
sterol regulatory element binding factor 2 |
88 |
0.94 |
| chr11_95723019_95723183 | 0.26 |
Zfp652 |
zinc finger protein 652 |
10090 |
0.12 |
| chr5_117361300_117361465 | 0.25 |
Wsb2 |
WD repeat and SOCS box-containing 2 |
360 |
0.77 |
| chr13_68712216_68712409 | 0.25 |
Gm26929 |
predicted gene, 26929 |
97768 |
0.06 |
| chr4_98923398_98923571 | 0.25 |
Usp1 |
ubiquitin specific peptidase 1 |
326 |
0.89 |
| chr11_52257389_52257568 | 0.25 |
Tcf7 |
transcription factor 7, T cell specific |
17399 |
0.11 |
| chr16_30925627_30925784 | 0.24 |
Gm46565 |
predicted gene, 46565 |
7635 |
0.17 |
| chr6_129254409_129254571 | 0.24 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
15765 |
0.11 |
| chr9_102864196_102864634 | 0.24 |
Ryk |
receptor-like tyrosine kinase |
4651 |
0.18 |
| chr4_8837559_8837760 | 0.24 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
2663 |
0.4 |
| chr10_80823975_80824142 | 0.24 |
Oaz1 |
ornithine decarboxylase antizyme 1 |
2598 |
0.1 |
| chr5_115491812_115492014 | 0.24 |
Gm13836 |
predicted gene 13836 |
542 |
0.51 |
| chr3_85264828_85265142 | 0.24 |
1700036G14Rik |
RIKEN cDNA 1700036G14 gene |
52534 |
0.14 |
| chr7_132554584_132554735 | 0.24 |
Oat |
ornithine aminotransferase |
21059 |
0.15 |
| chr17_33524094_33524279 | 0.24 |
Adamts10 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
18 |
0.97 |
| chr2_170826881_170827076 | 0.23 |
Dok5 |
docking protein 5 |
95171 |
0.08 |
| chr15_28106057_28106225 | 0.23 |
Gm31458 |
predicted gene, 31458 |
12311 |
0.17 |
| chr2_126520012_126520163 | 0.23 |
Atp8b4 |
ATPase, class I, type 8B, member 4 |
19413 |
0.19 |
| chr17_80728516_80728734 | 0.23 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
140 |
0.84 |
| chr1_85526452_85526618 | 0.23 |
AC147806.2 |
predicted 7592 |
67 |
0.59 |
| chr19_41829665_41829837 | 0.23 |
Frat1 |
frequently rearranged in advanced T cell lymphomas |
219 |
0.91 |
| chr7_104502188_104502379 | 0.23 |
Gm16464 |
predicted gene 16464 |
134 |
0.91 |
| chr8_126656365_126656543 | 0.23 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
62468 |
0.12 |
| chr7_126494300_126494471 | 0.23 |
Atxn2l |
ataxin 2-like |
1009 |
0.34 |
| chr1_85649860_85650030 | 0.23 |
Sp100 |
nuclear antigen Sp100 |
43 |
0.91 |
| chr4_60503881_60504482 | 0.22 |
Gm13773 |
predicted gene 13773 |
1340 |
0.3 |
| chr2_60963495_60963646 | 0.22 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
378 |
0.89 |
| chr19_25387114_25387309 | 0.22 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
8092 |
0.22 |
| chr6_146933231_146933413 | 0.22 |
1700034J05Rik |
RIKEN cDNA 1700034J05 gene |
21092 |
0.14 |
| chr17_86770158_86770337 | 0.22 |
Gm50008 |
predicted gene, 50008 |
3023 |
0.24 |
| chr5_52989351_52989502 | 0.22 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
2866 |
0.21 |
| chr2_84459790_84459967 | 0.22 |
Tfpi |
tissue factor pathway inhibitor |
1768 |
0.34 |
| chr8_94186443_94186638 | 0.22 |
Gm39228 |
predicted gene, 39228 |
3251 |
0.13 |
| chr11_116005461_116005624 | 0.22 |
Galk1 |
galactokinase 1 |
7177 |
0.1 |
| chr17_28257108_28257273 | 0.21 |
Ppard |
peroxisome proliferator activator receptor delta |
14518 |
0.11 |
| chr6_99171970_99172133 | 0.21 |
Foxp1 |
forkhead box P1 |
9033 |
0.29 |
| chr15_55819504_55819678 | 0.21 |
Sntb1 |
syntrophin, basic 1 |
86719 |
0.08 |
| chr11_49632419_49632570 | 0.21 |
Flt4 |
FMS-like tyrosine kinase 4 |
23231 |
0.1 |
| chr7_128740333_128740636 | 0.21 |
Mcmbp |
minichromosome maintenance complex binding protein |
2 |
0.95 |
| chr16_30067979_30068305 | 0.21 |
Hes1 |
hes family bHLH transcription factor 1 |
1804 |
0.29 |
| chr2_165855925_165856140 | 0.21 |
BC046401 |
cDNA sequence BC046401 |
1652 |
0.28 |
| chr11_70213843_70214036 | 0.21 |
Slc16a11 |
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
29 |
0.93 |
| chr10_75835778_75835936 | 0.21 |
Gstt2 |
glutathione S-transferase, theta 2 |
976 |
0.36 |
| chr1_85199424_85199611 | 0.21 |
AC123856.1 |
nuclear antigen Sp100 (Sp100) pseudogene |
96 |
0.49 |
| chr2_158179056_158179229 | 0.21 |
1700060C20Rik |
RIKEN cDNA 1700060C20 gene |
12866 |
0.14 |
| chr19_44997151_44997302 | 0.21 |
Sema4g |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
5090 |
0.11 |
| chr17_87474571_87474777 | 0.21 |
Gm30938 |
predicted gene, 30938 |
932 |
0.44 |
| chr1_86479051_86479408 | 0.21 |
Rpl30-ps6 |
ribosomal protein L30, pseudogene 6 |
5570 |
0.15 |
| chr5_121917596_121917747 | 0.21 |
Cux2 |
cut-like homeobox 2 |
937 |
0.54 |
| chr16_34745072_34745237 | 0.20 |
Mylk |
myosin, light polypeptide kinase |
56 |
0.98 |
| chr12_8774307_8774470 | 0.20 |
Sdc1 |
syndecan 1 |
2585 |
0.25 |
| chr7_25705726_25706061 | 0.20 |
Ccdc97 |
coiled-coil domain containing 97 |
10213 |
0.09 |
| chr8_70659044_70659249 | 0.20 |
Pgpep1 |
pyroglutamyl-peptidase I |
234 |
0.83 |
| chr6_52477529_52477702 | 0.20 |
1700094M24Rik |
RIKEN cDNA 1700094M24 gene |
14836 |
0.16 |
| chr12_84193897_84194204 | 0.20 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
35 |
0.95 |
| chr8_111501860_111502048 | 0.20 |
Wdr59 |
WD repeat domain 59 |
11201 |
0.17 |
| chr7_99355809_99355967 | 0.20 |
Serpinh1 |
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
2649 |
0.21 |
| chr7_15934182_15934342 | 0.20 |
Snord23 |
small nucleolar RNA, C/D box 23 |
4608 |
0.11 |
| chr1_85327350_85327504 | 0.20 |
Gm2619 |
predicted gene 2619 |
142 |
0.87 |
| chr4_58913611_58913779 | 0.20 |
Ecpas |
Ecm29 proteasome adaptor and scaffold |
946 |
0.53 |
| chr11_98777139_98777558 | 0.19 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2015 |
0.18 |
| chr7_141456726_141456885 | 0.19 |
Pnpla2 |
patatin-like phospholipase domain containing 2 |
213 |
0.8 |
| chr18_35814838_35814999 | 0.19 |
Ube2d2a |
ubiquitin-conjugating enzyme E2D 2A |
13611 |
0.09 |
| chr9_71165149_71165695 | 0.19 |
Aqp9 |
aquaporin 9 |
200 |
0.92 |
| chr1_135311434_135311608 | 0.19 |
Timm17a |
translocase of inner mitochondrial membrane 17a |
176 |
0.92 |
| chr5_63964660_63964830 | 0.19 |
Rell1 |
RELT-like 1 |
4078 |
0.19 |
| chr12_111428451_111428628 | 0.19 |
Exoc3l4 |
exocyst complex component 3-like 4 |
194 |
0.9 |
| chr8_120248017_120248168 | 0.19 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
19636 |
0.16 |
| chr3_95439810_95439972 | 0.19 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
5372 |
0.1 |
| chr8_22124952_22125137 | 0.19 |
Nek5 |
NIMA (never in mitosis gene a)-related expressed kinase 5 |
3 |
0.97 |
| chr13_75943581_75943755 | 0.19 |
Rhobtb3 |
Rho-related BTB domain containing 3 |
199 |
0.9 |
| chr9_48618793_48618948 | 0.19 |
Nnmt |
nicotinamide N-methyltransferase |
13717 |
0.24 |
| chr3_83953926_83954100 | 0.18 |
Tmem131l |
transmembrane 131 like |
10270 |
0.26 |
| chr13_30336361_30336758 | 0.18 |
Agtr1a |
angiotensin II receptor, type 1a |
115 |
0.97 |
| chr1_72827270_72827421 | 0.18 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
2023 |
0.37 |
| chr19_41346470_41346772 | 0.18 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
38475 |
0.17 |
| chr5_30816231_30816420 | 0.18 |
Gm19409 |
predicted gene, 19409 |
705 |
0.47 |
| chr1_58145718_58146241 | 0.18 |
Gm24548 |
predicted gene, 24548 |
9109 |
0.19 |
| chr8_46386589_46386784 | 0.18 |
Gm45253 |
predicted gene 45253 |
798 |
0.57 |
| chr7_19211436_19211587 | 0.18 |
Gpr4 |
G protein-coupled receptor 4 |
1027 |
0.3 |
| chr18_20945707_20945922 | 0.18 |
Rnf125 |
ring finger protein 125 |
1189 |
0.52 |
| chr14_103099863_103100150 | 0.18 |
Fbxl3 |
F-box and leucine-rich repeat protein 3 |
440 |
0.8 |
| chr12_25416673_25416856 | 0.18 |
Gm36723 |
predicted gene, 36723 |
22009 |
0.24 |
| chr11_120694977_120695348 | 0.18 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
5852 |
0.07 |
| chr4_108068074_108068398 | 0.17 |
Scp2 |
sterol carrier protein 2, liver |
3127 |
0.18 |
| chr15_79012870_79013120 | 0.17 |
Gm10865 |
predicted gene 10865 |
1954 |
0.13 |
| chr7_127805019_127805207 | 0.17 |
9430064I24Rik |
RIKEN cDNA 9430064I24 gene |
2351 |
0.11 |
| chr11_102051638_102051976 | 0.17 |
Cd300lg |
CD300 molecule like family member G |
10298 |
0.09 |
| chr3_151424962_151425113 | 0.17 |
Adgrl4 |
adhesion G protein-coupled receptor L4 |
12850 |
0.26 |
| chr14_61861512_61861706 | 0.17 |
Gm47952 |
predicted gene, 47952 |
59767 |
0.12 |
| chr1_191906223_191906394 | 0.17 |
Slc30a1 |
solute carrier family 30 (zinc transporter), member 1 |
459 |
0.69 |
| chr1_36556661_36556829 | 0.17 |
Sema4c |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
781 |
0.34 |
| chr6_88820995_88821174 | 0.17 |
Mgll |
monoglyceride lipase |
2046 |
0.19 |
| chr19_5663348_5663511 | 0.17 |
Sipa1 |
signal-induced proliferation associated gene 1 |
191 |
0.84 |
| chr11_106580051_106580208 | 0.17 |
Tex2 |
testis expressed gene 2 |
471 |
0.82 |
| chr8_119417331_119417482 | 0.17 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16718 |
0.14 |
| chr15_84714847_84715041 | 0.17 |
Arhgap8 |
Rho GTPase activating protein 8 |
5108 |
0.18 |
| chr9_94538161_94538340 | 0.17 |
Dipk2a |
divergent protein kinase domain 2A |
169 |
0.96 |
| chr9_37613490_37613650 | 0.17 |
Spa17 |
sperm autoantigenic protein 17 |
0 |
0.52 |
| chr9_107657905_107658551 | 0.17 |
Slc38a3 |
solute carrier family 38, member 3 |
967 |
0.31 |
| chr3_90537214_90537374 | 0.17 |
S100a16 |
S100 calcium binding protein A16 |
21 |
0.93 |
| chr18_20058884_20059060 | 0.17 |
Dsc2 |
desmocollin 2 |
506 |
0.84 |
| chr1_191254917_191255072 | 0.17 |
Gm37074 |
predicted gene, 37074 |
5570 |
0.14 |
| chr1_155050256_155050434 | 0.17 |
Gm29282 |
predicted gene 29282 |
6834 |
0.17 |
| chr15_25940308_25940999 | 0.17 |
Retreg1 |
reticulophagy regulator 1 |
49 |
0.98 |
| chrX_12072855_12073006 | 0.17 |
Bcor |
BCL6 interacting corepressor |
7623 |
0.27 |
| chr4_134995142_134995297 | 0.17 |
Syf2 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
64209 |
0.09 |
| chr12_84052558_84052962 | 0.17 |
Acot3 |
acyl-CoA thioesterase 3 |
336 |
0.69 |
| chr3_57344020_57344185 | 0.16 |
Gm5276 |
predicted gene 5276 |
18286 |
0.19 |
| chr2_167375199_167375534 | 0.16 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
26183 |
0.15 |
| chr11_95827586_95827744 | 0.16 |
Phospho1 |
phosphatase, orphan 1 |
3098 |
0.14 |
| chr11_113008652_113008818 | 0.16 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
164342 |
0.03 |
| chr8_125337571_125337768 | 0.16 |
Gm16237 |
predicted gene 16237 |
110346 |
0.06 |
| chr8_111776845_111777015 | 0.16 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
33121 |
0.14 |
| chr15_99716487_99716652 | 0.16 |
Gpd1 |
glycerol-3-phosphate dehydrogenase 1 (soluble) |
946 |
0.3 |
| chr3_85821987_85822152 | 0.16 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4778 |
0.2 |
| chr4_32520450_32520651 | 0.16 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
19045 |
0.19 |
| chr11_119045218_119045965 | 0.16 |
Cbx8 |
chromobox 8 |
4622 |
0.17 |
| chr2_30692293_30692476 | 0.16 |
Gm14486 |
predicted gene 14486 |
14373 |
0.14 |
| chr4_123412152_123412346 | 0.16 |
Macf1 |
microtubule-actin crosslinking factor 1 |
21 |
0.97 |
| chr10_4107762_4107973 | 0.16 |
Gm25515 |
predicted gene, 25515 |
3554 |
0.27 |
| chr4_106988955_106989270 | 0.16 |
Ssbp3 |
single-stranded DNA binding protein 3 |
22686 |
0.15 |
| chr11_97800088_97800264 | 0.16 |
Lasp1 |
LIM and SH3 protein 1 |
5 |
0.94 |
| chr5_125308383_125308534 | 0.16 |
Scarb1 |
scavenger receptor class B, member 1 |
3916 |
0.18 |
| chr10_118699777_118699944 | 0.16 |
Gm33337 |
predicted gene, 33337 |
15866 |
0.18 |
| chr15_83009671_83009841 | 0.16 |
Nfam1 |
Nfat activating molecule with ITAM motif 1 |
13327 |
0.13 |
| chr2_26578415_26578758 | 0.16 |
Egfl7 |
EGF-like domain 7 |
1428 |
0.23 |
| chr9_70318576_70318770 | 0.16 |
Mir5626 |
microRNA 5626 |
87080 |
0.07 |
| chr15_59717023_59717209 | 0.16 |
Gm20150 |
predicted gene, 20150 |
45003 |
0.15 |
| chr1_172312301_172312452 | 0.16 |
Igsf8 |
immunoglobulin superfamily, member 8 |
9 |
0.96 |
| chr12_102470243_102470394 | 0.16 |
Golga5 |
golgi autoantigen, golgin subfamily a, 5 |
1126 |
0.47 |
| chr10_20669582_20669754 | 0.15 |
Gm17230 |
predicted gene 17230 |
44033 |
0.15 |
| chr10_61313715_61314040 | 0.15 |
Rpl27a-ps1 |
ribosomal protein L27A, pseudogene 1 |
4047 |
0.16 |
| chr7_45782387_45782600 | 0.15 |
Lmtk3 |
lemur tyrosine kinase 3 |
1245 |
0.21 |
| chr4_124709066_124709229 | 0.15 |
Sf3a3 |
splicing factor 3a, subunit 3 |
5629 |
0.09 |
| chr2_24335745_24336056 | 0.15 |
Il1rn |
interleukin 1 receptor antagonist |
953 |
0.46 |
| chr10_80244946_80245115 | 0.15 |
Pwwp3a |
PWWP domain containing 3A, DNA repair factor |
2418 |
0.12 |
| chr12_108275649_108275819 | 0.15 |
Ccdc85c |
coiled-coil domain containing 85C |
309 |
0.89 |
| chr4_133556563_133556731 | 0.15 |
Gm23158 |
predicted gene, 23158 |
2302 |
0.15 |
| chr11_97435186_97435350 | 0.15 |
Arhgap23 |
Rho GTPase activating protein 23 |
1017 |
0.5 |
| chr9_32400631_32400981 | 0.15 |
Kcnj1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
6821 |
0.18 |
| chr16_35310487_35310877 | 0.15 |
Gm49706 |
predicted gene, 49706 |
12384 |
0.15 |
| chrX_13245137_13245293 | 0.15 |
2010308F09Rik |
RIKEN cDNA 2010308F09 gene |
2521 |
0.17 |
| chr7_25776793_25776952 | 0.15 |
Axl |
AXL receptor tyrosine kinase |
2194 |
0.16 |
| chr19_29142167_29142403 | 0.15 |
Mir101b |
microRNA 101b |
7006 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.3 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.2 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 0.2 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:1901321 | positive regulation of heart induction(GO:1901321) |
| 0.0 | 0.0 | GO:0052151 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:2000054 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.0 | GO:2000591 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.2 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |