Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
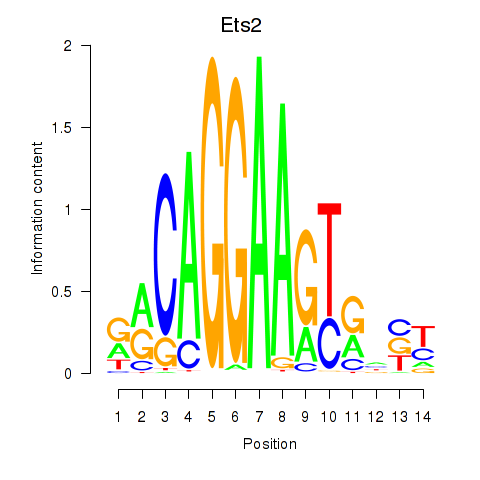
Results for Ets2
Z-value: 1.56
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSMUSG00000022895.8 | E26 avian leukemia oncogene 2, 3' domain |
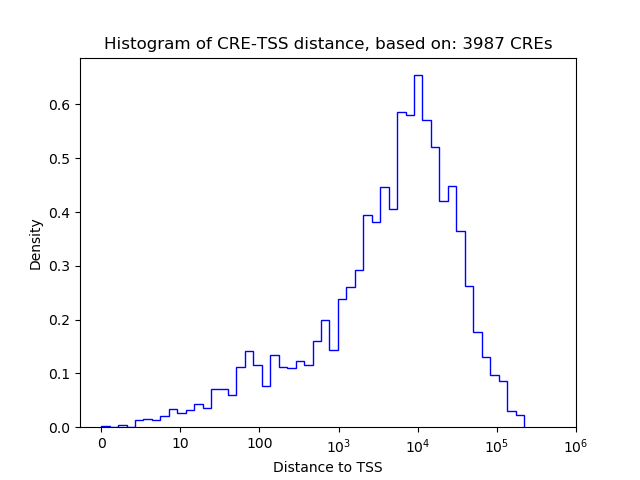
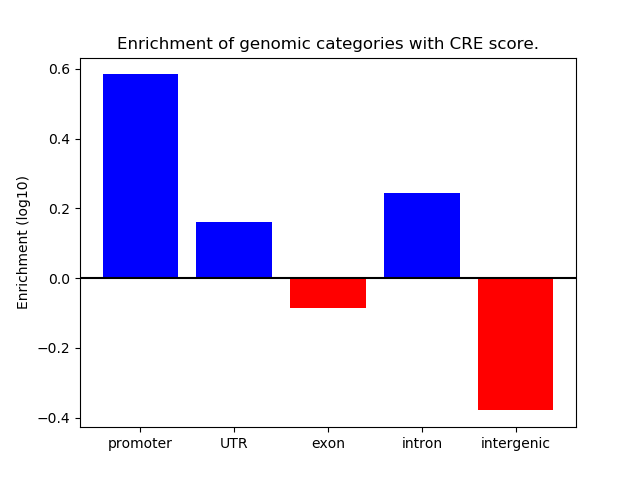
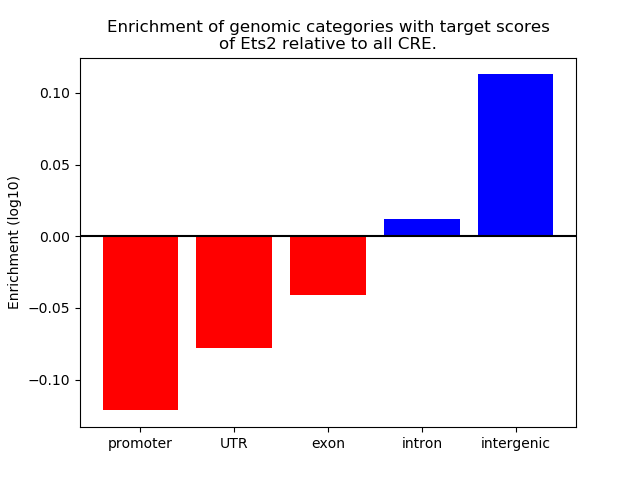
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_95700323_95700474 | Ets2 | 1677 | 0.411202 | 0.87 | 2.4e-02 | Click! |
| chr16_95674781_95674947 | Ets2 | 27211 | 0.189461 | -0.79 | 6.3e-02 | Click! |
| chr16_95682151_95682417 | Ets2 | 19791 | 0.204719 | 0.78 | 6.8e-02 | Click! |
| chr16_95702849_95703157 | Ets2 | 503 | 0.821559 | -0.78 | 6.9e-02 | Click! |
| chr16_95729140_95729334 | Ets2 | 11503 | 0.179276 | 0.76 | 8.2e-02 | Click! |
Activity of the Ets2 motif across conditions
Conditions sorted by the z-value of the Ets2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
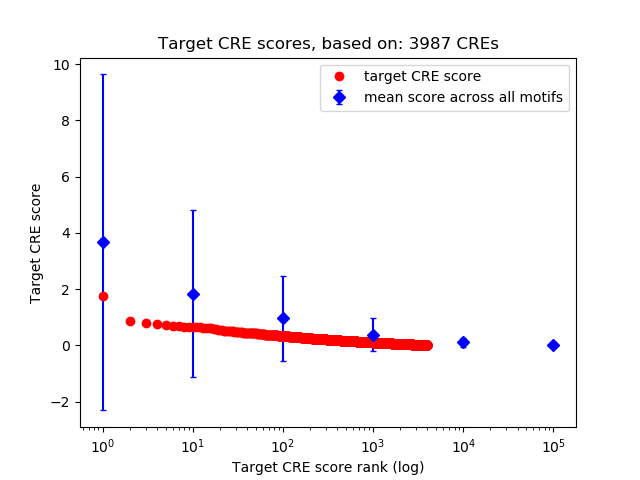
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_98767399_98767584 | 1.76 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2935 |
0.14 |
| chr3_103378119_103378272 | 0.86 |
Gm25009 |
predicted gene, 25009 |
14738 |
0.15 |
| chr2_164942950_164943143 | 0.79 |
Mmp9 |
matrix metallopeptidase 9 |
2266 |
0.19 |
| chr19_49421746_49421913 | 0.75 |
Gm50444 |
predicted gene, 50444 |
132189 |
0.06 |
| chr14_50981405_50981569 | 0.72 |
Gm8518 |
predicted gene 8518 |
2551 |
0.12 |
| chr17_88341719_88341880 | 0.69 |
Gm22542 |
predicted gene, 22542 |
683 |
0.69 |
| chr9_69028339_69028490 | 0.67 |
Rora |
RAR-related orphan receptor alpha |
167127 |
0.04 |
| chr11_88897701_88897852 | 0.66 |
Gm23507 |
predicted gene, 23507 |
7345 |
0.13 |
| chr14_7977646_7977810 | 0.66 |
Dnase1l3 |
deoxyribonuclease 1-like 3 |
506 |
0.78 |
| chr5_76063621_76063793 | 0.66 |
Gm6051 |
predicted gene 6051 |
2944 |
0.23 |
| chr17_36867279_36867493 | 0.65 |
Trim15 |
tripartite motif-containing 15 |
176 |
0.87 |
| chr3_55828967_55829118 | 0.64 |
4933417G07Rik |
RIKEN cDNA 4933417G07 gene |
18063 |
0.18 |
| chr1_119495139_119495303 | 0.63 |
Gm29454 |
predicted gene 29454 |
4256 |
0.15 |
| chr18_79032313_79032478 | 0.62 |
Setbp1 |
SET binding protein 1 |
76996 |
0.12 |
| chr15_57921642_57921803 | 0.61 |
Tbc1d31 |
TBC1 domain family, member 31 |
5709 |
0.21 |
| chr10_12278450_12278601 | 0.61 |
Gm48723 |
predicted gene, 48723 |
46136 |
0.14 |
| chr12_110381296_110381622 | 0.59 |
Gm47195 |
predicted gene, 47195 |
56074 |
0.08 |
| chr15_28106057_28106225 | 0.57 |
Gm31458 |
predicted gene, 31458 |
12311 |
0.17 |
| chr3_145834049_145834216 | 0.56 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
23653 |
0.18 |
| chr11_78267972_78268123 | 0.56 |
2610507B11Rik |
RIKEN cDNA 2610507B11 gene |
1914 |
0.15 |
| chr4_11084820_11085206 | 0.55 |
Ndufaf6 |
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
8808 |
0.15 |
| chr2_60922422_60922578 | 0.52 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
33470 |
0.19 |
| chr1_151597907_151598058 | 0.52 |
Fam129a |
family with sequence similarity 129, member A |
26486 |
0.15 |
| chr2_125263901_125264052 | 0.51 |
A530010F05Rik |
RIKEN cDNA A530010F05 gene |
10588 |
0.16 |
| chr4_138440154_138440340 | 0.51 |
Mul1 |
mitochondrial ubiquitin ligase activator of NFKB 1 |
5524 |
0.14 |
| chr11_84062884_84063069 | 0.51 |
Dusp14 |
dual specificity phosphatase 14 |
4104 |
0.2 |
| chr4_127263896_127264076 | 0.50 |
Smim12 |
small integral membrane protein 12 |
20202 |
0.12 |
| chr14_79157797_79157989 | 0.49 |
Gm49015 |
predicted gene, 49015 |
6504 |
0.16 |
| chr10_62506271_62506458 | 0.49 |
Srgn |
serglycin |
1391 |
0.34 |
| chr18_38190654_38190820 | 0.48 |
Pcdh1 |
protocadherin 1 |
12426 |
0.13 |
| chr19_11977402_11977711 | 0.48 |
Osbp |
oxysterol binding protein |
11615 |
0.09 |
| chr19_42610535_42610705 | 0.47 |
Loxl4 |
lysyl oxidase-like 4 |
2089 |
0.31 |
| chr1_23279889_23280199 | 0.47 |
Mir30a |
microRNA 30a |
7775 |
0.12 |
| chr9_74328886_74329063 | 0.46 |
Gm24141 |
predicted gene, 24141 |
33636 |
0.17 |
| chr7_132735250_132735442 | 0.46 |
Fam53b |
family with sequence similarity 53, member B |
41570 |
0.13 |
| chr19_55491305_55491575 | 0.45 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
110426 |
0.07 |
| chr10_85354450_85354601 | 0.45 |
Gm25281 |
predicted gene, 25281 |
20624 |
0.16 |
| chr18_83740581_83740741 | 0.44 |
Gm50419 |
predicted gene, 50419 |
86945 |
0.08 |
| chr11_16737408_16737579 | 0.44 |
Gm25698 |
predicted gene, 25698 |
4782 |
0.22 |
| chr3_146846305_146846456 | 0.44 |
Ttll7 |
tubulin tyrosine ligase-like family, member 7 |
5987 |
0.18 |
| chr16_95449403_95449694 | 0.44 |
Erg |
ETS transcription factor |
9697 |
0.27 |
| chr1_39710733_39710905 | 0.43 |
Rfx8 |
regulatory factor X 8 |
10178 |
0.19 |
| chr11_6478757_6478908 | 0.43 |
Purb |
purine rich element binding protein B |
2915 |
0.11 |
| chr5_117361300_117361465 | 0.43 |
Wsb2 |
WD repeat and SOCS box-containing 2 |
360 |
0.77 |
| chr14_63133716_63133871 | 0.43 |
Ctsb |
cathepsin B |
1725 |
0.28 |
| chr14_22269831_22270192 | 0.43 |
Lrmda |
leucine rich melanocyte differentiation associated |
40154 |
0.2 |
| chr11_103223555_103223709 | 0.43 |
Map3k14 |
mitogen-activated protein kinase kinase kinase 14 |
1785 |
0.22 |
| chr4_105257759_105257936 | 0.43 |
Plpp3 |
phospholipid phosphatase 3 |
100500 |
0.08 |
| chr1_38109860_38110024 | 0.42 |
Rev1 |
REV1, DNA directed polymerase |
18359 |
0.15 |
| chr7_71704948_71705119 | 0.42 |
Gm44688 |
predicted gene 44688 |
1043 |
0.49 |
| chr3_127355382_127355546 | 0.42 |
Gm42969 |
predicted gene 42969 |
38242 |
0.11 |
| chr7_80013937_80014119 | 0.42 |
Zfp710 |
zinc finger protein 710 |
10786 |
0.12 |
| chr9_54844002_54844168 | 0.42 |
Rab7-ps1 |
RAB7, member RAS oncogene family, pseudogene 1 |
18803 |
0.15 |
| chr1_186584026_186584190 | 0.41 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
25427 |
0.19 |
| chr17_8128409_8128574 | 0.41 |
Rnaset2a |
ribonuclease T2A |
19294 |
0.13 |
| chr6_88584043_88584194 | 0.40 |
Kbtbd12 |
kelch repeat and BTB (POZ) domain containing 12 |
6541 |
0.19 |
| chr17_6998212_6998376 | 0.40 |
Rnaset2b |
ribonuclease T2B |
7638 |
0.13 |
| chr11_4261437_4261588 | 0.40 |
Lif |
leukemia inhibitory factor |
3955 |
0.13 |
| chr6_116004145_116004296 | 0.40 |
Plxnd1 |
plexin D1 |
9215 |
0.16 |
| chr13_111984044_111984208 | 0.39 |
Gm15322 |
predicted gene 15322 |
7340 |
0.2 |
| chr8_105504683_105504849 | 0.39 |
Gm8838 |
predicted gene 8838 |
1360 |
0.25 |
| chr16_84972074_84972233 | 0.39 |
Gm10791 |
predicted gene 10791 |
61 |
0.97 |
| chr2_167831188_167831379 | 0.38 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
2363 |
0.25 |
| chr15_97590730_97591033 | 0.38 |
Gm49506 |
predicted gene, 49506 |
32591 |
0.19 |
| chr11_121686812_121686981 | 0.38 |
B3gntl1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
13743 |
0.18 |
| chrY_90828861_90829019 | 0.37 |
Gm21742 |
predicted gene, 21742 |
8473 |
0.19 |
| chr9_71606651_71607004 | 0.37 |
Gm6018 |
predicted gene 6018 |
6687 |
0.19 |
| chr3_95312699_95312852 | 0.37 |
Cers2 |
ceramide synthase 2 |
2016 |
0.14 |
| chr16_32130242_32130457 | 0.37 |
Nrros |
negative regulator of reactive oxygen species |
17532 |
0.1 |
| chr5_105698740_105698909 | 0.37 |
Lrrc8d |
leucine rich repeat containing 8D |
1145 |
0.57 |
| chr6_72473587_72473785 | 0.37 |
Mat2a |
methionine adenosyltransferase II, alpha |
34128 |
0.09 |
| chr11_83224315_83224478 | 0.36 |
Gm11427 |
predicted gene 11427 |
58 |
0.73 |
| chr8_88983561_88983712 | 0.36 |
Mir8110 |
microRNA 8110 |
41099 |
0.17 |
| chr2_50824872_50825029 | 0.36 |
Gm13498 |
predicted gene 13498 |
84734 |
0.1 |
| chr6_72663055_72663233 | 0.36 |
Tcf7l1 |
transcription factor 7 like 1 (T cell specific, HMG box) |
8260 |
0.1 |
| chr4_104840212_104840384 | 0.35 |
C8a |
complement component 8, alpha polypeptide |
16225 |
0.18 |
| chr8_84068735_84068886 | 0.35 |
C330011M18Rik |
RIKEN cDNA C330011M18 gene |
1523 |
0.16 |
| chr8_122313304_122313653 | 0.35 |
Zfpm1 |
zinc finger protein, multitype 1 |
6158 |
0.15 |
| chr2_152359606_152359999 | 0.35 |
Gm14165 |
predicted gene 14165 |
6187 |
0.1 |
| chr12_5213695_5213866 | 0.35 |
Gm48532 |
predicted gene, 48532 |
22184 |
0.23 |
| chr5_139329080_139329231 | 0.35 |
Adap1 |
ArfGAP with dual PH domains 1 |
3533 |
0.16 |
| chr5_25100632_25101023 | 0.35 |
2900005J15Rik |
RIKEN cDNA 2900005J15 gene |
156 |
0.53 |
| chr7_126494300_126494471 | 0.35 |
Atxn2l |
ataxin 2-like |
1009 |
0.34 |
| chr15_27925949_27926114 | 0.35 |
Trio |
triple functional domain (PTPRF interacting) |
6760 |
0.24 |
| chr19_5274259_5274410 | 0.35 |
Pacs1 |
phosphofurin acidic cluster sorting protein 1 |
1215 |
0.24 |
| chr4_133169582_133169763 | 0.35 |
Wasf2 |
WAS protein family, member 2 |
6329 |
0.15 |
| chr11_117878325_117878508 | 0.35 |
Tha1 |
threonine aldolase 1 |
4935 |
0.11 |
| chr2_31825447_31825664 | 0.35 |
Qrfp |
pyroglutamylated RFamide peptide |
14975 |
0.13 |
| chr11_3289571_3289722 | 0.35 |
Patz1 |
POZ (BTB) and AT hook containing zinc finger 1 |
347 |
0.8 |
| chr9_124423659_124423821 | 0.34 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
157 |
0.94 |
| chr16_84834907_84835088 | 0.34 |
Atp5j |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
8 |
0.54 |
| chr11_79223575_79223745 | 0.34 |
Gm11200 |
predicted gene 11200 |
10864 |
0.17 |
| chr11_80067662_80067813 | 0.34 |
Crlf3 |
cytokine receptor-like factor 3 |
9838 |
0.16 |
| chr15_53724232_53724383 | 0.33 |
1700015H07Rik |
RIKEN cDNA 1700015H07 gene |
102652 |
0.07 |
| chr10_121521092_121521243 | 0.33 |
Gm35696 |
predicted gene, 35696 |
10769 |
0.11 |
| chr7_145102326_145102499 | 0.33 |
Gm45181 |
predicted gene 45181 |
60584 |
0.12 |
| chr5_134572591_134572794 | 0.33 |
Mir7228 |
microRNA 7228 |
616 |
0.56 |
| chr11_16844158_16844466 | 0.33 |
Egfros |
epidermal growth factor receptor, opposite strand |
13610 |
0.2 |
| chr3_137860459_137860760 | 0.33 |
Gm43403 |
predicted gene 43403 |
3574 |
0.12 |
| chr9_109118225_109118412 | 0.33 |
Plxnb1 |
plexin B1 |
11524 |
0.1 |
| chr10_78135134_78135285 | 0.33 |
Gm47922 |
predicted gene, 47922 |
2888 |
0.18 |
| chr15_77343141_77343292 | 0.33 |
Gm49436 |
predicted gene, 49436 |
23483 |
0.1 |
| chr17_48039561_48039744 | 0.33 |
1700122O11Rik |
RIKEN cDNA 1700122O11 gene |
1359 |
0.33 |
| chr15_73153357_73153508 | 0.33 |
Ago2 |
argonaute RISC catalytic subunit 2 |
24415 |
0.17 |
| chr11_106382531_106382850 | 0.33 |
Icam2 |
intercellular adhesion molecule 2 |
91 |
0.96 |
| chr10_72778559_72778735 | 0.33 |
Gm47094 |
predicted gene, 47094 |
60716 |
0.14 |
| chr10_75048271_75048422 | 0.32 |
Rab36 |
RAB36, member RAS oncogene family |
10123 |
0.15 |
| chr2_104817028_104817755 | 0.32 |
Qser1 |
glutamine and serine rich 1 |
631 |
0.64 |
| chr7_25776793_25776952 | 0.32 |
Axl |
AXL receptor tyrosine kinase |
2194 |
0.16 |
| chr8_120041013_120041184 | 0.32 |
Gm15684 |
predicted gene 15684 |
4189 |
0.17 |
| chr16_30066871_30067038 | 0.32 |
Hes1 |
hes family bHLH transcription factor 1 |
616 |
0.68 |
| chr11_115641190_115641341 | 0.32 |
Slc25a19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
12970 |
0.09 |
| chr15_99333031_99333200 | 0.32 |
Fmnl3 |
formin-like 3 |
2612 |
0.15 |
| chr15_64043505_64043715 | 0.32 |
Fam49b |
family with sequence similarity 49, member B |
16720 |
0.16 |
| chr4_41760316_41760485 | 0.31 |
Il11ra1 |
interleukin 11 receptor, alpha chain 1 |
54 |
0.94 |
| chr1_191154426_191154595 | 0.31 |
Fam71a |
family with sequence similarity 71, member A |
10307 |
0.13 |
| chr13_5592555_5592743 | 0.31 |
Gm35330 |
predicted gene, 35330 |
21521 |
0.26 |
| chr6_142708472_142708687 | 0.31 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
6264 |
0.22 |
| chr13_119723468_119723673 | 0.31 |
Nim1k |
NIM1 serine/threonine protein kinase |
14824 |
0.11 |
| chr16_49801576_49801764 | 0.31 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
1137 |
0.53 |
| chr7_101360514_101360680 | 0.31 |
Arap1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
665 |
0.54 |
| chr13_5845761_5845936 | 0.31 |
1700016G22Rik |
RIKEN cDNA 1700016G22 gene |
11713 |
0.16 |
| chr2_179583110_179583261 | 0.31 |
Gm14300 |
predicted gene 14300 |
30382 |
0.23 |
| chr18_46253564_46253715 | 0.31 |
Pggt1b |
protein geranylgeranyltransferase type I, beta subunit |
4734 |
0.19 |
| chr5_32745803_32745961 | 0.31 |
Pisd |
phosphatidylserine decarboxylase |
430 |
0.74 |
| chr16_44029399_44029560 | 0.31 |
Gramd1c |
GRAM domain containing 1C |
1534 |
0.37 |
| chr12_69516624_69516775 | 0.31 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
41149 |
0.1 |
| chr4_136090324_136090475 | 0.30 |
Gm13009 |
predicted gene 13009 |
15427 |
0.12 |
| chr10_107361652_107361839 | 0.30 |
Gm38117 |
predicted gene, 38117 |
75614 |
0.09 |
| chr19_34199620_34199771 | 0.30 |
Stambpl1 |
STAM binding protein like 1 |
6873 |
0.18 |
| chr1_165617212_165617387 | 0.30 |
Mpzl1 |
myelin protein zero-like 1 |
3320 |
0.14 |
| chr11_16895683_16895855 | 0.30 |
Egfr |
epidermal growth factor receptor |
9416 |
0.2 |
| chr8_126757967_126758167 | 0.30 |
Gm45805 |
predicted gene 45805 |
267 |
0.94 |
| chr3_35318331_35318524 | 0.30 |
Gm25442 |
predicted gene, 25442 |
7187 |
0.25 |
| chr9_16115179_16115336 | 0.30 |
Fat3 |
FAT atypical cadherin 3 |
108507 |
0.07 |
| chr10_93285572_93285740 | 0.30 |
Elk3 |
ELK3, member of ETS oncogene family |
25155 |
0.14 |
| chr16_11006083_11006392 | 0.30 |
Gm24961 |
predicted gene, 24961 |
11851 |
0.1 |
| chr14_63957868_63958033 | 0.29 |
Sox7 |
SRY (sex determining region Y)-box 7 |
14277 |
0.16 |
| chr17_58480733_58480887 | 0.29 |
Gm49848 |
predicted gene, 49848 |
18485 |
0.25 |
| chr5_53214446_53214609 | 0.29 |
Sel1l3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
600 |
0.76 |
| chr17_9512595_9512746 | 0.29 |
Gm49807 |
predicted gene, 49807 |
37021 |
0.18 |
| chr11_78600166_78600339 | 0.29 |
Gm11196 |
predicted gene 11196 |
14697 |
0.1 |
| chr11_16756467_16757088 | 0.29 |
Egfr |
epidermal growth factor receptor |
4547 |
0.23 |
| chr14_73838973_73839138 | 0.29 |
Gm16409 |
predicted gene 16409 |
41765 |
0.13 |
| chr3_142745096_142745348 | 0.29 |
Gm15540 |
predicted gene 15540 |
541 |
0.67 |
| chr12_73715979_73716146 | 0.29 |
Prkch |
protein kinase C, eta |
67141 |
0.1 |
| chr9_45247764_45247948 | 0.28 |
Gm20612 |
predicted gene 20612 |
13199 |
0.1 |
| chr9_118611265_118611450 | 0.28 |
Itga9 |
integrin alpha 9 |
4667 |
0.19 |
| chr18_21242831_21243130 | 0.28 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
57143 |
0.11 |
| chr6_117431300_117431501 | 0.28 |
Gm4640 |
predicted gene 4640 |
7783 |
0.22 |
| chr9_79605873_79606044 | 0.28 |
Col12a1 |
collagen, type XII, alpha 1 |
7012 |
0.21 |
| chr9_110326292_110326443 | 0.28 |
Gm42638 |
predicted gene 42638 |
4197 |
0.13 |
| chr19_37248362_37248513 | 0.28 |
Gm25268 |
predicted gene, 25268 |
14953 |
0.13 |
| chr9_23011584_23011737 | 0.28 |
Bmper |
BMP-binding endothelial regulator |
211416 |
0.02 |
| chr10_21946855_21947006 | 0.28 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
18460 |
0.17 |
| chr13_18800559_18800710 | 0.28 |
Vps41 |
VPS41 HOPS complex subunit |
20791 |
0.24 |
| chr17_28029449_28029600 | 0.28 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
11251 |
0.12 |
| chr5_124074959_124075110 | 0.28 |
Abcb9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
4097 |
0.12 |
| chr17_43242727_43242878 | 0.28 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
27527 |
0.22 |
| chr9_32901306_32901512 | 0.28 |
Gm27162 |
predicted gene 27162 |
27557 |
0.18 |
| chr14_65113058_65113209 | 0.28 |
Extl3 |
exostosin-like glycosyltransferase 3 |
15027 |
0.18 |
| chr8_83773624_83773779 | 0.28 |
Gm45778 |
predicted gene 45778 |
24197 |
0.11 |
| chr8_119424899_119425070 | 0.27 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
9140 |
0.15 |
| chr8_117321338_117321500 | 0.27 |
Cmip |
c-Maf inducing protein |
27751 |
0.21 |
| chr15_10981382_10981708 | 0.27 |
Amacr |
alpha-methylacyl-CoA racemase |
211 |
0.92 |
| chr1_85526452_85526618 | 0.27 |
AC147806.2 |
predicted 7592 |
67 |
0.59 |
| chr2_69116662_69116813 | 0.27 |
Nostrin |
nitric oxide synthase trafficker |
19063 |
0.19 |
| chr4_117124519_117125009 | 0.27 |
Btbd19 |
BTB (POZ) domain containing 19 |
402 |
0.61 |
| chr1_93634572_93634756 | 0.27 |
Gm28536 |
predicted gene 28536 |
529 |
0.56 |
| chr2_18944090_18944247 | 0.27 |
Pip4k2a |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
37894 |
0.17 |
| chr10_61929391_61929572 | 0.27 |
Gm5750 |
predicted gene 5750 |
30143 |
0.16 |
| chr16_90677368_90677525 | 0.27 |
Gm36169 |
predicted gene, 36169 |
446 |
0.78 |
| chr10_116264669_116264820 | 0.27 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
10779 |
0.19 |
| chr11_16844975_16845160 | 0.27 |
Egfros |
epidermal growth factor receptor, opposite strand |
14365 |
0.2 |
| chr2_28526560_28526711 | 0.27 |
Ralgds |
ral guanine nucleotide dissociation stimulator |
1215 |
0.31 |
| chr15_77606069_77606360 | 0.26 |
Gm36245 |
predicted gene, 36245 |
6956 |
0.11 |
| chr5_35729958_35730139 | 0.26 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
772 |
0.61 |
| chr6_142649490_142649655 | 0.26 |
5330439B14Rik |
RIKEN cDNA 5330439B14 gene |
34977 |
0.15 |
| chr13_111933651_111933850 | 0.26 |
Gm9025 |
predicted gene 9025 |
49573 |
0.1 |
| chr15_64345201_64345377 | 0.26 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
4597 |
0.21 |
| chr10_94565775_94566229 | 0.26 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
9255 |
0.17 |
| chr1_150459711_150460052 | 0.26 |
Prg4 |
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
5717 |
0.2 |
| chr9_29666708_29666884 | 0.26 |
Gm15521 |
predicted gene 15521 |
74286 |
0.13 |
| chr1_125489382_125489583 | 0.26 |
Gm28706 |
predicted gene 28706 |
44915 |
0.15 |
| chr10_78326387_78326551 | 0.26 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
5779 |
0.09 |
| chr12_35021403_35021554 | 0.25 |
Snx13 |
sorting nexin 13 |
25708 |
0.16 |
| chr16_33580716_33580902 | 0.25 |
Slc12a8 |
solute carrier family 12 (potassium/chloride transporters), member 8 |
15753 |
0.25 |
| chr19_10236152_10236493 | 0.25 |
Myrf |
myelin regulatory factor |
4282 |
0.13 |
| chr7_49178293_49178474 | 0.25 |
Gm37613 |
predicted gene, 37613 |
17158 |
0.18 |
| chr13_111896407_111896568 | 0.25 |
Gm9025 |
predicted gene 9025 |
12310 |
0.15 |
| chr16_4512630_4512833 | 0.25 |
Srl |
sarcalumenin |
10332 |
0.16 |
| chr15_77626960_77627111 | 0.25 |
Gm8221 |
predicted gene 8221 |
5201 |
0.11 |
| chr1_85649860_85650030 | 0.25 |
Sp100 |
nuclear antigen Sp100 |
43 |
0.91 |
| chr9_32906562_32906941 | 0.25 |
Gm27162 |
predicted gene 27162 |
22215 |
0.19 |
| chr6_142346265_142346455 | 0.25 |
Pyroxd1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
576 |
0.69 |
| chr2_71867523_71867680 | 0.25 |
Pdk1 |
pyruvate dehydrogenase kinase, isoenzyme 1 |
5623 |
0.2 |
| chr12_80887036_80887400 | 0.25 |
Susd6 |
sushi domain containing 6 |
18510 |
0.13 |
| chr11_57445339_57445531 | 0.25 |
Gm12242 |
predicted gene 12242 |
3672 |
0.29 |
| chr10_25434450_25434623 | 0.25 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
1920 |
0.35 |
| chr13_112116146_112116297 | 0.25 |
Gm31104 |
predicted gene, 31104 |
21895 |
0.19 |
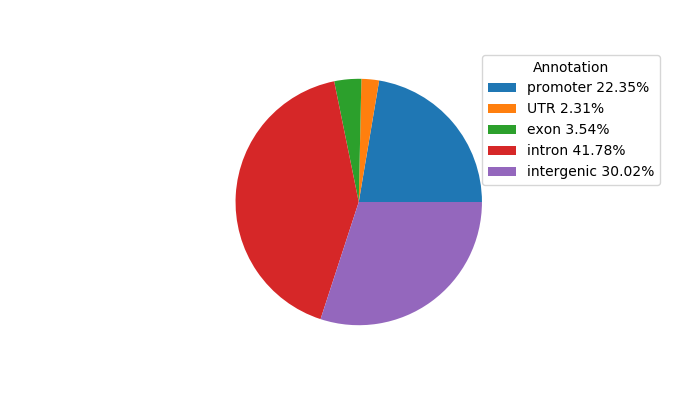
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.2 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.3 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0045402 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0044805 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0072711 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0046719 | regulation by virus of viral protein levels in host cell(GO:0046719) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.2 | GO:0032610 | interleukin-1 alpha production(GO:0032610) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.0 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0060956 | endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.0 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.0 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.0 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |