Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
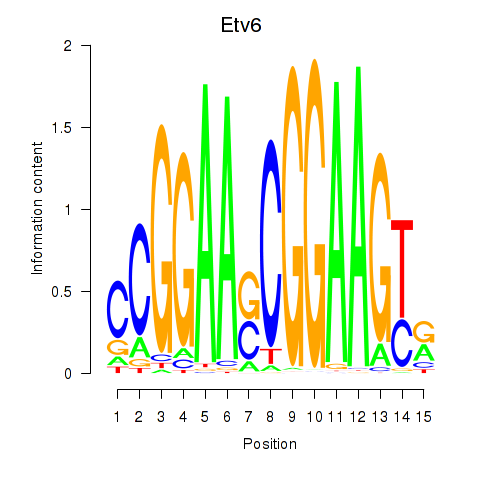
Results for Etv6
Z-value: 1.60
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.10 | ets variant 6 |
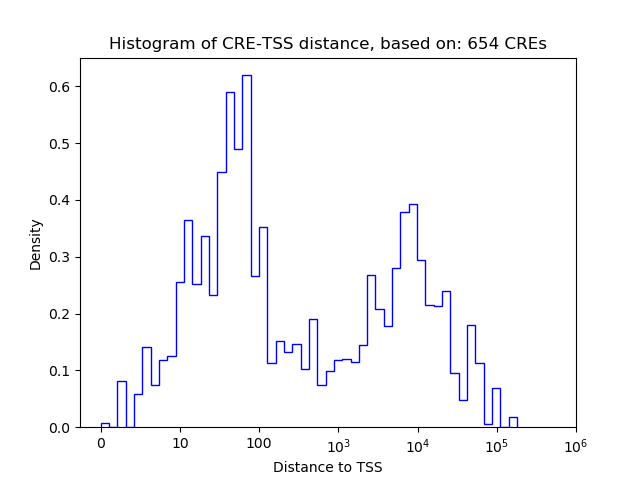
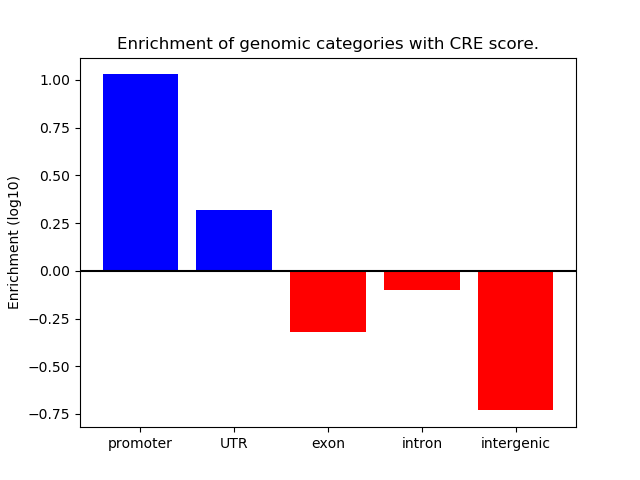
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_134039206_134039407 | Etv6 | 3337 | 0.250610 | -0.97 | 1.2e-03 | Click! |
| chr6_134271340_134271530 | Etv6 | 23870 | 0.149971 | 0.91 | 1.2e-02 | Click! |
| chr6_134036525_134036733 | Etv6 | 660 | 0.723229 | -0.89 | 1.8e-02 | Click! |
| chr6_134036923_134037340 | Etv6 | 1162 | 0.507613 | -0.87 | 2.6e-02 | Click! |
| chr6_134045191_134045355 | Etv6 | 9304 | 0.190962 | 0.84 | 3.7e-02 | Click! |
Activity of the Etv6 motif across conditions
Conditions sorted by the z-value of the Etv6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
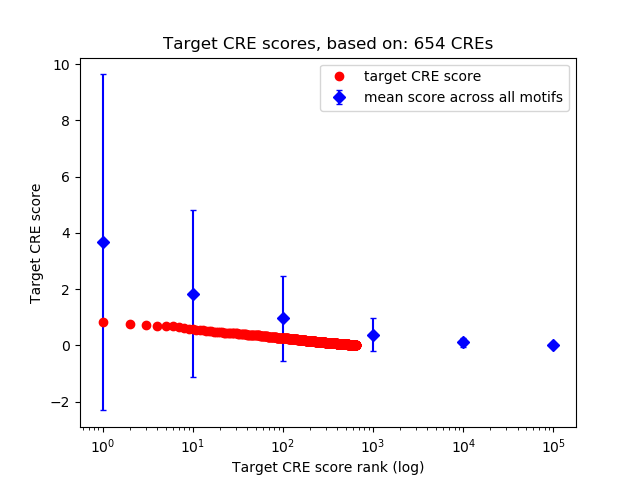
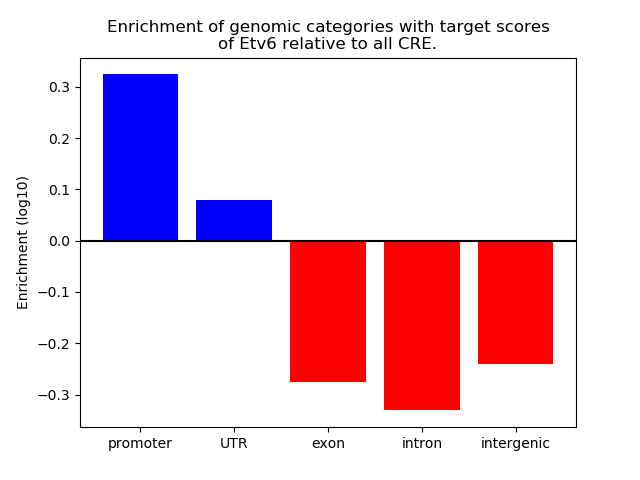
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_156523955_156524106 | 0.84 |
Csnk2a3 |
casein kinase 2 alpha 3 |
18 |
0.97 |
| chr14_20319722_20320198 | 0.75 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr9_45677619_45677770 | 0.71 |
Dscaml1 |
DS cell adhesion molecule like 1 |
4857 |
0.22 |
| chr5_116024413_116024572 | 0.68 |
Prkab1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
16 |
0.96 |
| chr14_20315223_20315409 | 0.67 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
3887 |
0.15 |
| chrX_60547467_60547618 | 0.67 |
Gm715 |
predicted gene 715 |
477 |
0.75 |
| chr1_171418992_171419158 | 0.65 |
Tstd1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
42 |
0.93 |
| chr7_4792782_4792944 | 0.62 |
Rpl28 |
ribosomal protein L28 |
11 |
0.93 |
| chr10_58255342_58255504 | 0.57 |
Gcc2 |
GRIP and coiled-coil domain containing 2 |
74 |
0.96 |
| chr4_136886108_136886460 | 0.57 |
C1qb |
complement component 1, q subcomponent, beta polypeptide |
97 |
0.96 |
| chr8_105297519_105297695 | 0.53 |
E2f4 |
E2F transcription factor 4 |
56 |
0.9 |
| chr5_33658354_33658511 | 0.53 |
Tmem129 |
transmembrane protein 129 |
16 |
0.6 |
| chr11_85492360_85492531 | 0.53 |
Bcas3 |
breast carcinoma amplified sequence 3 |
123 |
0.95 |
| chr10_116264669_116264820 | 0.52 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
10779 |
0.19 |
| chr7_48917526_48917677 | 0.52 |
Gm45282 |
predicted gene 45282 |
7539 |
0.14 |
| chr10_75834373_75834559 | 0.50 |
Gstt2 |
glutathione S-transferase, theta 2 |
3 |
0.95 |
| chr2_27540118_27540305 | 0.49 |
Gm13421 |
predicted gene 13421 |
215 |
0.57 |
| chr11_88204256_88204424 | 0.49 |
Mrps23 |
mitochondrial ribosomal protein S23 |
48 |
0.97 |
| chr2_3727636_3727853 | 0.48 |
Fam107b |
family with sequence similarity 107, member B |
14266 |
0.16 |
| chr11_5485683_5486034 | 0.47 |
Gm11962 |
predicted gene 11962 |
9588 |
0.14 |
| chr9_107563062_107563244 | 0.46 |
Tusc2 |
tumor suppressor 2, mitochondrial calcium regulator |
102 |
0.87 |
| chr16_89830199_89830569 | 0.45 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
7491 |
0.3 |
| chr5_125361058_125361209 | 0.45 |
Gm40323 |
predicted gene, 40323 |
42 |
0.96 |
| chr11_69554847_69555031 | 0.45 |
Efnb3 |
ephrin B3 |
5266 |
0.08 |
| chr2_94010737_94010899 | 0.44 |
Alkbh3 |
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
11 |
0.97 |
| chr6_120822682_120822842 | 0.44 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
31 |
0.97 |
| chr3_109255070_109255263 | 0.44 |
Gm13865 |
predicted gene 13865 |
62299 |
0.11 |
| chr13_21362713_21362885 | 0.43 |
Zscan12 |
zinc finger and SCAN domain containing 12 |
21 |
0.94 |
| chr5_105112436_105112587 | 0.43 |
Gbp9 |
guanylate-binding protein 9 |
2234 |
0.28 |
| chr8_11009560_11009740 | 0.42 |
Irs2 |
insulin receptor substrate 2 |
1192 |
0.34 |
| chr17_20945145_20945356 | 0.42 |
Ppp2r1a |
protein phosphatase 2, regulatory subunit A, alpha |
61 |
0.54 |
| chr1_133610283_133610442 | 0.41 |
Snrpe |
small nuclear ribonucleoprotein E |
71 |
0.96 |
| chr5_3490465_3490637 | 0.41 |
Gm42435 |
predicted gene 42435 |
9521 |
0.13 |
| chr1_180226517_180226708 | 0.41 |
Psen2 |
presenilin 2 |
2956 |
0.19 |
| chr17_27839932_27840089 | 0.41 |
Snrpc |
U1 small nuclear ribonucleoprotein C |
74 |
0.95 |
| chr2_58516954_58517105 | 0.40 |
Acvr1 |
activin A receptor, type 1 |
757 |
0.71 |
| chr11_86757488_86757692 | 0.40 |
Cltc |
clathrin, heavy polypeptide (Hc) |
25 |
0.98 |
| chr10_128805402_128805563 | 0.40 |
Dnajc14 |
DnaJ heat shock protein family (Hsp40) member C14 |
72 |
0.91 |
| chr15_79670588_79670885 | 0.39 |
Tomm22 |
translocase of outer mitochondrial membrane 22 |
125 |
0.91 |
| chr7_105744339_105744512 | 0.38 |
Taf10 |
TATA-box binding protein associated factor 10 |
64 |
0.94 |
| chr10_13008300_13008462 | 0.38 |
Sf3b5 |
splicing factor 3b, subunit 5 |
3038 |
0.25 |
| chr8_94183485_94183637 | 0.38 |
Gm39228 |
predicted gene, 39228 |
272 |
0.83 |
| chr7_99483674_99483841 | 0.38 |
Rps3 |
ribosomal protein S3 |
19 |
0.93 |
| chr10_59629065_59629266 | 0.37 |
Mcu |
mitochondrial calcium uniporter |
12473 |
0.19 |
| chr9_65032284_65032455 | 0.37 |
Dpp8 |
dipeptidylpeptidase 8 |
45 |
0.97 |
| chr10_10457121_10457304 | 0.37 |
Gm48326 |
predicted gene, 48326 |
8322 |
0.2 |
| chr11_70764308_70764498 | 0.36 |
Zfp3 |
zinc finger protein 3 |
194 |
0.85 |
| chr11_5955636_5955797 | 0.36 |
Ykt6 |
YKT6 v-SNARE homolog (S. cerevisiae) |
23 |
0.97 |
| chr2_31052576_31052733 | 0.36 |
Fnbp1 |
formin binding protein 1 |
2790 |
0.24 |
| chr1_97770025_97770211 | 0.36 |
Gin1 |
gypsy retrotransposon integrase 1 |
54 |
0.8 |
| chr1_74289444_74289611 | 0.36 |
Gm37503 |
predicted gene, 37503 |
1667 |
0.15 |
| chr4_95354053_95354288 | 0.36 |
Gm29064 |
predicted gene 29064 |
48620 |
0.15 |
| chr14_51884664_51884952 | 0.36 |
Mettl17 |
methyltransferase like 17 |
34 |
0.94 |
| chr9_20581222_20581545 | 0.35 |
Zfp846 |
zinc finger protein 846 |
58 |
0.96 |
| chr11_3452291_3452451 | 0.35 |
8430429K09Rik |
RIKEN cDNA 8430429K09 gene |
6 |
0.49 |
| chr5_145201795_145201971 | 0.35 |
Zkscan14 |
zinc finger with KRAB and SCAN domains 14 |
15 |
0.95 |
| chr16_3872258_3872624 | 0.35 |
Zfp597 |
zinc finger protein 597 |
45 |
0.51 |
| chr9_110887920_110888088 | 0.35 |
Als2cl |
ALS2 C-terminal like |
9 |
0.95 |
| chr19_42631725_42631876 | 0.35 |
Loxl4 |
lysyl oxidase-like 4 |
18987 |
0.18 |
| chr2_94406648_94406807 | 0.34 |
Ttc17 |
tetratricopeptide repeat domain 17 |
38 |
0.91 |
| chr1_167308570_167308761 | 0.34 |
Tmco1 |
transmembrane and coiled-coil domains 1 |
5 |
0.96 |
| chr13_52041479_52041668 | 0.33 |
Gm37872 |
predicted gene, 37872 |
19646 |
0.19 |
| chr18_67448711_67448888 | 0.33 |
Afg3l2 |
AFG3-like AAA ATPase 2 |
308 |
0.86 |
| chr11_78245899_78246053 | 0.32 |
Supt6 |
SPT6, histone chaperone and transcription elongation factor |
11 |
0.73 |
| chr9_46272980_46273140 | 0.32 |
Zpr1 |
ZPR1 zinc finger |
4 |
0.94 |
| chr2_32535355_32535730 | 0.32 |
Fam102a |
family with sequence similarity 102, member A |
183 |
0.89 |
| chr19_45006439_45006606 | 0.32 |
Twnk |
twinkle mtDNA helicase |
36 |
0.53 |
| chr8_77610687_77610852 | 0.31 |
Tmem184c |
transmembrane protein 184C |
71 |
0.52 |
| chr14_122104924_122105095 | 0.31 |
A330035P11Rik |
RIKEN cDNA A330035P11 gene |
1944 |
0.21 |
| chr15_8109190_8109363 | 0.31 |
Nup155 |
nucleoporin 155 |
3 |
0.98 |
| chr19_28963790_28963966 | 0.30 |
4430402I18Rik |
RIKEN cDNA 4430402I18 gene |
30 |
0.54 |
| chr8_33885650_33885854 | 0.30 |
Gm26978 |
predicted gene, 26978 |
5 |
0.97 |
| chr11_77793325_77793488 | 0.30 |
Gm10277 |
predicted gene 10277 |
5659 |
0.16 |
| chr17_33948663_33948832 | 0.30 |
B3galt4 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
2741 |
0.06 |
| chr7_99694503_99694673 | 0.30 |
Gm34280 |
predicted gene, 34280 |
291 |
0.76 |
| chr9_20521327_20521505 | 0.30 |
Zfp266 |
zinc finger protein 266 |
1 |
0.96 |
| chr19_53332014_53332179 | 0.29 |
Mxi1 |
MAX interactor 1, dimerization protein |
1688 |
0.28 |
| chr9_20952862_20953022 | 0.29 |
Dnmt1 |
DNA methyltransferase (cytosine-5) 1 |
37 |
0.95 |
| chr17_36271362_36271517 | 0.29 |
Trim39 |
tripartite motif-containing 39 |
56 |
0.91 |
| chr2_27626035_27626261 | 0.29 |
Rxra |
retinoid X receptor alpha |
50292 |
0.12 |
| chr9_124424078_124424238 | 0.28 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
4 |
0.97 |
| chr15_34443209_34443373 | 0.28 |
Rpl30 |
ribosomal protein L30 |
32 |
0.8 |
| chr3_95217667_95217848 | 0.28 |
Gabpb2 |
GA repeat binding protein, beta 2 |
3 |
0.79 |
| chr13_64398164_64398509 | 0.28 |
Ctsl |
cathepsin L |
27446 |
0.09 |
| chr10_14705498_14705660 | 0.28 |
Nmbr |
neuromedin B receptor |
12 |
0.5 |
| chr4_125066548_125066718 | 0.27 |
Snip1 |
Smad nuclear interacting protein 1 |
39 |
0.93 |
| chr5_146770278_146770441 | 0.27 |
Usp12 |
ubiquitin specific peptidase 12 |
11903 |
0.15 |
| chr4_124802426_124802586 | 0.27 |
Mtf1 |
metal response element binding transcription factor 1 |
43 |
0.95 |
| chr4_133941503_133941734 | 0.27 |
Hmgn2 |
high mobility group nucleosomal binding domain 2 |
25625 |
0.11 |
| chr4_48279558_48279880 | 0.27 |
Invs |
inversin |
41 |
0.66 |
| chr17_26113206_26113368 | 0.27 |
Tmem8 |
transmembrane protein 8 |
12 |
0.94 |
| chr1_182520133_182520312 | 0.27 |
Capn2 |
calpain 2 |
2614 |
0.21 |
| chr12_108792186_108792364 | 0.27 |
Gm34220 |
predicted gene, 34220 |
468 |
0.53 |
| chr3_95871479_95871654 | 0.27 |
C920021L13Rik |
RIKEN cDNA C920021L13 gene |
35 |
0.49 |
| chr6_125009567_125009728 | 0.27 |
Zfp384 |
zinc finger protein 384 |
22 |
0.94 |
| chr4_81766890_81767041 | 0.27 |
Gm11412 |
predicted gene 11412 |
23819 |
0.23 |
| chr15_64060280_64060431 | 0.26 |
Fam49b |
family with sequence similarity 49, member B |
12 |
0.98 |
| chr2_121473990_121474147 | 0.26 |
Mfap1b |
microfibrillar-associated protein 1B |
1 |
0.94 |
| chr2_29935294_29935462 | 0.26 |
Gle1 |
GLE1 RNA export mediator (yeast) |
48 |
0.95 |
| chr15_66865965_66866140 | 0.26 |
Ccn4 |
cellular communication network factor 4 |
25268 |
0.16 |
| chr1_192770564_192770748 | 0.26 |
Hhat |
hedgehog acyltransferase |
336 |
0.87 |
| chr11_95146239_95146409 | 0.26 |
Dlx4 |
distal-less homeobox 4 |
61 |
0.95 |
| chr17_35241635_35241803 | 0.25 |
Ddx39b |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
27 |
0.88 |
| chr16_31948445_31948606 | 0.25 |
Ncbp2 |
nuclear cap binding protein subunit 2 |
12 |
0.49 |
| chr11_77078417_77078807 | 0.25 |
Nsrp1 |
nuclear speckle regulatory protein 1 |
177 |
0.75 |
| chr6_35539812_35539979 | 0.25 |
Mtpn |
myotrophin |
7 |
0.98 |
| chr13_46669359_46669525 | 0.25 |
Fam8a1 |
family with sequence similarity 8, member A1 |
80 |
0.97 |
| chr17_46621404_46621583 | 0.25 |
Ptk7 |
PTK7 protein tyrosine kinase 7 |
8011 |
0.1 |
| chr4_120975511_120975682 | 0.25 |
Smap2 |
small ArfGAP 2 |
112 |
0.94 |
| chr11_94653678_94653838 | 0.25 |
Mrpl27 |
mitochondrial ribosomal protein L27 |
9 |
0.56 |
| chr17_31305538_31305732 | 0.25 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
92 |
0.96 |
| chr17_8310765_8311153 | 0.25 |
4930506C21Rik |
RIKEN cDNA 4930506C21 gene |
93 |
0.41 |
| chr14_26669821_26669989 | 0.25 |
Pde12 |
phosphodiesterase 12 |
22 |
0.52 |
| chr11_83298893_83299082 | 0.24 |
Pex12 |
peroxisomal biogenesis factor 12 |
10 |
0.5 |
| chr16_16526849_16527032 | 0.24 |
Fgd4 |
FYVE, RhoGEF and PH domain containing 4 |
471 |
0.82 |
| chr7_79228544_79228706 | 0.24 |
Gm31510 |
predicted gene, 31510 |
44186 |
0.1 |
| chr1_167353970_167354146 | 0.24 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
3746 |
0.15 |
| chr17_49458493_49458669 | 0.24 |
Mocs1 |
molybdenum cofactor synthesis 1 |
6297 |
0.24 |
| chr6_126107373_126107694 | 0.24 |
Ntf3 |
neurotrophin 3 |
57427 |
0.15 |
| chr12_55155236_55155394 | 0.24 |
Srp54b |
signal recognition particle 54B |
55 |
0.85 |
| chr7_45434250_45434440 | 0.24 |
Ruvbl2 |
RuvB-like protein 2 |
68 |
0.83 |
| chr10_81606036_81606372 | 0.23 |
BC025920 |
cDNA sequence BC025920 |
66 |
0.93 |
| chr15_98506392_98506556 | 0.23 |
Olfr278-ps1 |
olfactory receptor 278, pseudogene 1 |
7069 |
0.07 |
| chr3_108256959_108257142 | 0.23 |
1700010K24Rik |
RIKEN cDNA 1700010K24 gene |
65 |
0.56 |
| chr5_9087032_9087225 | 0.23 |
Tmem243 |
transmembrane protein 243, mitochondrial |
13540 |
0.16 |
| chr2_91412402_91412624 | 0.23 |
Gm22071 |
predicted gene, 22071 |
5377 |
0.18 |
| chr7_45923080_45923254 | 0.23 |
Ccdc114 |
coiled-coil domain containing 114 |
905 |
0.25 |
| chr2_145934675_145934834 | 0.23 |
Cfap61 |
cilia and flagella associated protein 61 |
30 |
0.5 |
| chr12_55080388_55080552 | 0.23 |
Srp54a |
signal recognition particle 54A |
56 |
0.82 |
| chr5_105789979_105790426 | 0.23 |
Rps15a-ps5 |
ribosomal protein S15A, pseudogene 5 |
7511 |
0.17 |
| chr8_120324733_120324902 | 0.22 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
96361 |
0.06 |
| chrX_168673452_168673623 | 0.22 |
Msl3 |
MSL complex subunit 3 |
361 |
0.91 |
| chr8_84856734_84857027 | 0.22 |
Farsa |
phenylalanyl-tRNA synthetase, alpha subunit |
109 |
0.9 |
| chr7_28796738_28796906 | 0.22 |
Rinl |
Ras and Rab interactor-like |
116 |
0.91 |
| chr3_101524513_101524690 | 0.22 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
52959 |
0.11 |
| chr1_36244254_36244418 | 0.22 |
Uggt1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
31 |
0.97 |
| chr9_78489041_78489208 | 0.22 |
Eef1a1 |
eukaryotic translation elongation factor 1 alpha 1 |
27 |
0.96 |
| chr6_52160159_52160321 | 0.22 |
Hotairm1 |
Hoxa transcript antisense RNA, myeloid-specific 1 |
1716 |
0.12 |
| chr8_10857974_10858150 | 0.22 |
Gm32540 |
predicted gene, 32540 |
8124 |
0.12 |
| chr11_46039470_46039639 | 0.22 |
Gm12165 |
predicted gene 12165 |
3114 |
0.18 |
| chr19_5366591_5366746 | 0.21 |
Banf1 |
BAF nuclear assembly factor 1 |
62 |
0.49 |
| chr2_75978121_75978296 | 0.21 |
Ttc30a2 |
tetratricopeptide repeat domain 30A2 |
38 |
0.97 |
| chr2_48814064_48814223 | 0.21 |
Acvr2a |
activin receptor IIA |
34 |
0.98 |
| chr11_48800219_48800381 | 0.21 |
Rack1 |
receptor for activated C kinase 1 |
32 |
0.88 |
| chr17_26663048_26663210 | 0.20 |
Atp6v0e |
ATPase, H+ transporting, lysosomal V0 subunit E |
102 |
0.96 |
| chr19_8770923_8771084 | 0.20 |
Tmem223 |
transmembrane protein 223 |
7 |
0.87 |
| chr9_41105205_41105372 | 0.20 |
Gm48611 |
predicted gene, 48611 |
6793 |
0.17 |
| chr8_69808918_69809151 | 0.20 |
Gmip |
Gem-interacting protein |
344 |
0.77 |
| chr16_91740939_91741112 | 0.20 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
10791 |
0.14 |
| chrX_7841056_7841418 | 0.20 |
Otud5 |
OTU domain containing 5 |
127 |
0.91 |
| chr17_33915764_33916103 | 0.20 |
Zbtb22 |
zinc finger and BTB domain containing 22 |
29 |
0.48 |
| chrX_12376263_12376437 | 0.20 |
Gm14635 |
predicted gene 14635 |
21354 |
0.24 |
| chr10_80129893_80130054 | 0.20 |
Stk11 |
serine/threonine kinase 11 |
3291 |
0.11 |
| chr5_122318215_122318386 | 0.20 |
Gm15842 |
predicted gene 15842 |
15878 |
0.1 |
| chr11_78436121_78436295 | 0.20 |
Gm11195 |
predicted gene 11195 |
7899 |
0.09 |
| chr2_19004905_19005068 | 0.20 |
4930426L09Rik |
RIKEN cDNA 4930426L09 gene |
6643 |
0.22 |
| chr1_118321721_118321879 | 0.19 |
Nifk |
nucleolar protein interacting with the FHA domain of MKI67 |
39 |
0.97 |
| chr9_20951552_20951703 | 0.19 |
Dnmt1 |
DNA methyltransferase (cytosine-5) 1 |
978 |
0.34 |
| chr1_171113920_171114112 | 0.19 |
Cfap126 |
cilia and flagella associated protein 126 |
27 |
0.95 |
| chr17_26916833_26917013 | 0.19 |
Kifc5b |
kinesin family member C5B |
168 |
0.87 |
| chr13_17993285_17993442 | 0.19 |
Yae1d1 |
Yae1 domain containing 1 |
14 |
0.93 |
| chr9_43210448_43210605 | 0.19 |
Pou2f3 |
POU domain, class 2, transcription factor 3 |
157 |
0.94 |
| chr9_37348340_37348499 | 0.18 |
Ccdc15 |
coiled-coil domain containing 15 |
13 |
0.96 |
| chr7_126695761_126695919 | 0.18 |
Slx1b |
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
56 |
0.57 |
| chr12_25239865_25240032 | 0.18 |
Gm19340 |
predicted gene, 19340 |
42647 |
0.13 |
| chr19_46136477_46136807 | 0.18 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
647 |
0.58 |
| chr6_124965441_124965599 | 0.18 |
Cops7a |
COP9 signalosome subunit 7A |
8 |
0.93 |
| chr2_71039832_71040019 | 0.18 |
Dcaf17 |
DDB1 and CUL4 associated factor 17 |
15403 |
0.19 |
| chr5_143758154_143758320 | 0.18 |
D130017N08Rik |
RIKEN cDNA D130017N08 gene |
117 |
0.96 |
| chr2_84714853_84715028 | 0.18 |
Zdhhc5 |
zinc finger, DHHC domain containing 5 |
96 |
0.92 |
| chr6_48708145_48708369 | 0.18 |
Gimap6 |
GTPase, IMAP family member 6 |
32 |
0.93 |
| chr3_107850765_107850942 | 0.18 |
Gm36211 |
predicted gene, 36211 |
17314 |
0.08 |
| chr9_49486024_49486190 | 0.18 |
Ttc12 |
tetratricopeptide repeat domain 12 |
108 |
0.97 |
| chr10_80320493_80320652 | 0.18 |
2310011J03Rik |
RIKEN cDNA 2310011J03 gene |
35 |
0.92 |
| chr6_91877905_91878069 | 0.18 |
Gm44712 |
predicted gene 44712 |
2 |
0.54 |
| chr11_58330593_58330753 | 0.17 |
Sh3bp5l |
SH3 binding domain protein 5 like |
51 |
0.81 |
| chr10_22731356_22731520 | 0.17 |
Tbpl1 |
TATA box binding protein-like 1 |
9 |
0.72 |
| chr6_92042166_92042321 | 0.17 |
Fgd5 |
FYVE, RhoGEF and PH domain containing 5 |
25186 |
0.13 |
| chr13_52024582_52024974 | 0.17 |
Gm37872 |
predicted gene, 37872 |
2851 |
0.3 |
| chr18_60848929_60849091 | 0.17 |
Tcof1 |
treacle ribosome biogenesis factor 1 |
39 |
0.97 |
| chr2_60878102_60878265 | 0.17 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
3255 |
0.35 |
| chr10_80151775_80151933 | 0.17 |
Midn |
midnolin |
616 |
0.5 |
| chr10_128547738_128547893 | 0.17 |
Zc3h10 |
zinc finger CCCH type containing 10 |
41 |
0.91 |
| chr5_100663119_100663274 | 0.17 |
Coq2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
1223 |
0.4 |
| chr12_7946863_7947014 | 0.17 |
Gm48633 |
predicted gene, 48633 |
3123 |
0.25 |
| chr10_67285195_67285355 | 0.17 |
Nrbf2 |
nuclear receptor binding factor 2 |
30 |
0.97 |
| chr4_44065513_44065687 | 0.17 |
Gne |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
1320 |
0.35 |
| chr2_26493017_26493193 | 0.16 |
Notch1 |
notch 1 |
10717 |
0.1 |
| chr13_56588544_56588882 | 0.16 |
2010203P06Rik |
RIKEN cDNA 2010203P06 gene |
6824 |
0.18 |
| chr1_132362082_132362267 | 0.16 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
2774 |
0.18 |
| chr11_4849530_4849684 | 0.16 |
Nf2 |
neurofibromin 2 |
71 |
0.97 |
| chr11_84867701_84867877 | 0.16 |
Ggnbp2 |
gametogenetin binding protein 2 |
2413 |
0.18 |
| chr11_73137165_73137468 | 0.16 |
Haspin |
histone H3 associated protein kinase |
978 |
0.41 |
| chr2_90904641_90904800 | 0.16 |
Ndufs3 |
NADH:ubiquinone oxidoreductase core subunit S3 |
18 |
0.49 |
| chr11_16508132_16508289 | 0.16 |
Sec61g |
SEC61, gamma subunit |
44 |
0.98 |
| chr7_128237495_128237652 | 0.16 |
Armc5 |
armadillo repeat containing 5 |
216 |
0.67 |
| chr11_9373284_9373435 | 0.16 |
Abca13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
179908 |
0.03 |
| chrX_107816272_107816433 | 0.16 |
2610002M06Rik |
RIKEN cDNA 2610002M06 gene |
18 |
0.62 |
| chr16_25052954_25053153 | 0.16 |
A230028O05Rik |
RIKEN cDNA A230028O05 gene |
6586 |
0.32 |
| chr3_94886891_94887048 | 0.16 |
Psmb4 |
proteasome (prosome, macropain) subunit, beta type 4 |
8 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.4 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.4 | GO:0052173 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.1 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.4 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.1 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.2 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.7 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006056 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |