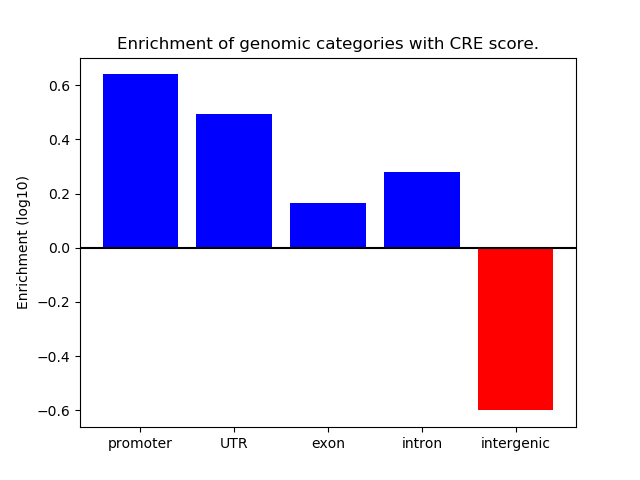
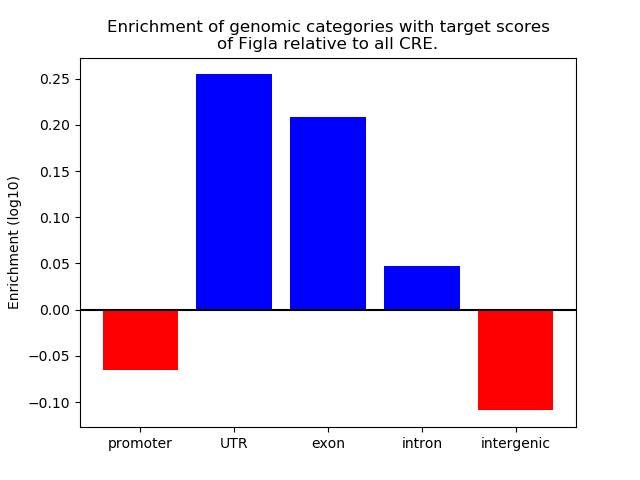
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
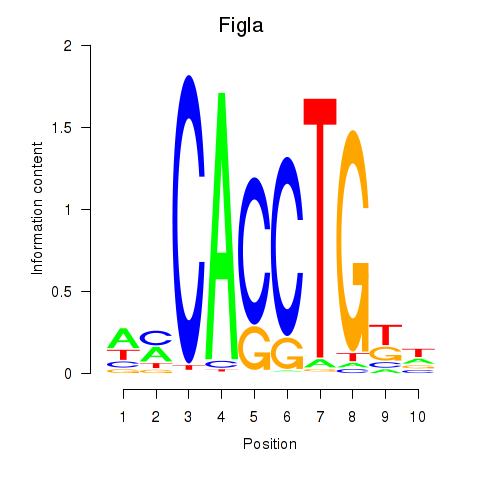
Results for Figla
Z-value: 0.78
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSMUSG00000030001.3 | folliculogenesis specific basic helix-loop-helix |
Activity of the Figla motif across conditions
Conditions sorted by the z-value of the Figla motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
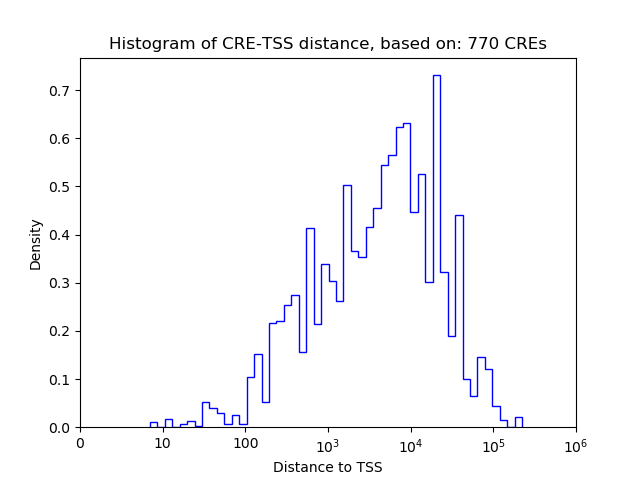
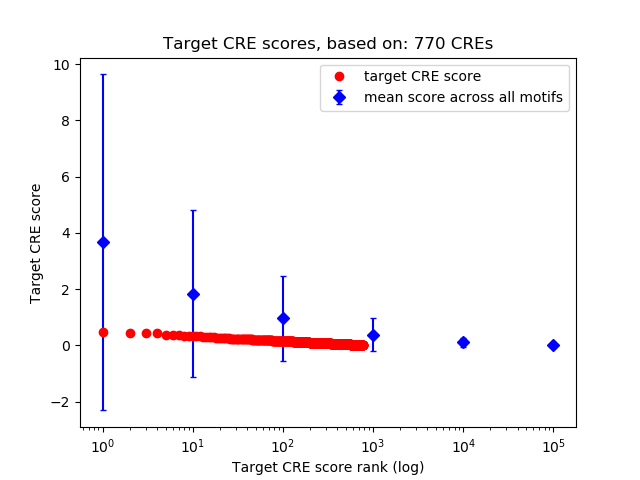
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_51255928_51256079 | 0.47 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr1_67146253_67146467 | 0.45 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
23334 |
0.21 |
| chr2_84995821_84996178 | 0.45 |
Prg3 |
proteoglycan 3 |
7784 |
0.12 |
| chr2_32525017_32525309 | 0.44 |
Gm13412 |
predicted gene 13412 |
132 |
0.92 |
| chr19_44404842_44405075 | 0.38 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
1732 |
0.29 |
| chr1_60501628_60501779 | 0.37 |
Raph1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
1562 |
0.34 |
| chr11_110380321_110380711 | 0.35 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
18606 |
0.23 |
| chr1_51770461_51770858 | 0.34 |
Myo1b |
myosin IB |
220 |
0.94 |
| chr15_88866027_88866463 | 0.33 |
Pim3 |
proviral integration site 3 |
1325 |
0.34 |
| chr2_70813245_70813396 | 0.32 |
Tlk1 |
tousled-like kinase 1 |
11908 |
0.21 |
| chr3_97651056_97651341 | 0.32 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
6995 |
0.14 |
| chr19_58385888_58386039 | 0.32 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
68503 |
0.11 |
| chr5_125040097_125040498 | 0.31 |
Ncor2 |
nuclear receptor co-repressor 2 |
2515 |
0.25 |
| chr9_21671190_21671387 | 0.31 |
Smarca4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
35521 |
0.09 |
| chr10_89451030_89451181 | 0.29 |
Gas2l3 |
growth arrest-specific 2 like 3 |
7138 |
0.25 |
| chr15_59059061_59059543 | 0.29 |
Mtss1 |
MTSS I-BAR domain containing 1 |
3838 |
0.27 |
| chr14_11132816_11133186 | 0.29 |
Fhit |
fragile histidine triad gene |
7410 |
0.24 |
| chr11_78845894_78846146 | 0.28 |
Lyrm9 |
LYR motif containing 9 |
19391 |
0.15 |
| chr4_49490545_49490860 | 0.28 |
Baat |
bile acid-Coenzyme A: amino acid N-acyltransferase |
12418 |
0.12 |
| chr4_53218925_53219106 | 0.27 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
1158 |
0.48 |
| chr1_127899545_127899941 | 0.27 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1170 |
0.47 |
| chr4_88889115_88889274 | 0.27 |
Ifne |
interferon epsilon |
8993 |
0.08 |
| chr19_25542114_25542287 | 0.27 |
Dmrt1 |
doublesex and mab-3 related transcription factor 1 |
36493 |
0.15 |
| chr5_125492454_125492640 | 0.26 |
Gm27551 |
predicted gene, 27551 |
13170 |
0.13 |
| chr10_68131050_68131381 | 0.24 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
5411 |
0.28 |
| chr2_155052449_155052605 | 0.24 |
a |
nonagouti |
4912 |
0.16 |
| chr4_45470057_45470208 | 0.24 |
Shb |
src homology 2 domain-containing transforming protein B |
12670 |
0.16 |
| chr15_3742307_3742458 | 0.24 |
Gm4823 |
predicted gene 4823 |
4493 |
0.32 |
| chr8_40881703_40881854 | 0.24 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
6830 |
0.17 |
| chr3_51185339_51185556 | 0.23 |
Noct |
nocturnin |
39000 |
0.12 |
| chr11_11844763_11844947 | 0.23 |
Ddc |
dopa decarboxylase |
8575 |
0.18 |
| chr9_120936662_120936813 | 0.23 |
Ctnnb1 |
catenin (cadherin associated protein), beta 1 |
2919 |
0.15 |
| chr13_111488300_111488451 | 0.22 |
Gpbp1 |
GC-rich promoter binding protein 1 |
663 |
0.7 |
| chr1_92901975_92902158 | 0.22 |
Ankmy1 |
ankyrin repeat and MYND domain containing 1 |
840 |
0.41 |
| chr9_123021710_123021910 | 0.22 |
Gm9856 |
predicted gene 9856 |
238 |
0.48 |
| chr8_61273697_61273901 | 0.22 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
15073 |
0.2 |
| chr1_16271768_16272085 | 0.22 |
Gm7568 |
predicted gene 7568 |
18297 |
0.19 |
| chr5_28463981_28464172 | 0.21 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
2908 |
0.25 |
| chr4_53121067_53121218 | 0.21 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
38753 |
0.15 |
| chr9_94053316_94053467 | 0.21 |
Gm5369 |
predicted gene 5369 |
86071 |
0.11 |
| chr8_68051671_68051822 | 0.21 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
10481 |
0.24 |
| chr5_135693560_135693734 | 0.21 |
Por |
P450 (cytochrome) oxidoreductase |
4489 |
0.12 |
| chr7_39809957_39810110 | 0.21 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
12291 |
0.16 |
| chr16_93819391_93819545 | 0.21 |
Dop1b |
DOP1 leucine zipper like protein B |
10034 |
0.13 |
| chr15_57710526_57710885 | 0.21 |
Gm16006 |
predicted gene 16006 |
1778 |
0.36 |
| chr2_181461827_181461978 | 0.21 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
2476 |
0.17 |
| chr19_40223143_40223322 | 0.20 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
124 |
0.95 |
| chr6_119266532_119266713 | 0.20 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
916 |
0.63 |
| chr8_12805253_12805404 | 0.20 |
Atp11a |
ATPase, class VI, type 11A |
6088 |
0.17 |
| chr1_23290878_23291029 | 0.20 |
Gm27028 |
predicted gene, 27028 |
584 |
0.46 |
| chr10_62590569_62590741 | 0.20 |
Ddx21 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 |
316 |
0.82 |
| chr8_122935865_122936016 | 0.20 |
Ankrd11 |
ankyrin repeat domain 11 |
19953 |
0.13 |
| chr7_25452202_25452383 | 0.20 |
Gm15495 |
predicted gene 15495 |
8829 |
0.1 |
| chr4_132489447_132489680 | 0.20 |
Gm12981 |
predicted gene 12981 |
20086 |
0.09 |
| chr11_35693250_35693421 | 0.20 |
Pank3 |
pantothenate kinase 3 |
76149 |
0.08 |
| chr5_125070237_125070401 | 0.19 |
Ncor2 |
nuclear receptor co-repressor 2 |
11908 |
0.16 |
| chr12_104079953_104080165 | 0.19 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
590 |
0.59 |
| chr8_47532977_47533306 | 0.19 |
Trappc11 |
trafficking protein particle complex 11 |
326 |
0.63 |
| chr5_122455814_122455965 | 0.19 |
Anapc7 |
anaphase promoting complex subunit 7 |
14514 |
0.09 |
| chr8_84842789_84843130 | 0.19 |
Calr |
calreticulin |
1957 |
0.13 |
| chr14_120500061_120500218 | 0.19 |
Rap2a |
RAS related protein 2a |
21695 |
0.23 |
| chr10_69584120_69584461 | 0.19 |
Gm46231 |
predicted gene, 46231 |
17929 |
0.24 |
| chr4_45464623_45464804 | 0.19 |
Shb |
src homology 2 domain-containing transforming protein B |
18089 |
0.15 |
| chr2_31478552_31479205 | 0.18 |
Ass1 |
argininosuccinate synthetase 1 |
8671 |
0.19 |
| chr14_21088324_21088564 | 0.18 |
Adk |
adenosine kinase |
12292 |
0.21 |
| chr13_113765532_113765689 | 0.18 |
Gm47463 |
predicted gene, 47463 |
995 |
0.38 |
| chr1_186682239_186682607 | 0.18 |
Gm26169 |
predicted gene, 26169 |
5681 |
0.16 |
| chr9_35026486_35026660 | 0.18 |
Kirrel3 |
kirre like nephrin family adhesion molecule 3 |
10121 |
0.18 |
| chr6_11931201_11931441 | 0.18 |
Phf14 |
PHD finger protein 14 |
4538 |
0.29 |
| chr10_128397387_128397714 | 0.18 |
Gm17201 |
predicted gene 17201 |
1987 |
0.11 |
| chr2_181251704_181251904 | 0.17 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
4327 |
0.11 |
| chr18_61809305_61809458 | 0.17 |
Afap1l1 |
actin filament associated protein 1-like 1 |
22679 |
0.16 |
| chr3_51416743_51417219 | 0.17 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
367 |
0.75 |
| chr19_20610582_20610763 | 0.17 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
8711 |
0.22 |
| chr12_16585344_16585551 | 0.17 |
Lpin1 |
lipin 1 |
4273 |
0.27 |
| chr7_99173709_99174071 | 0.17 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
3784 |
0.16 |
| chr8_22875512_22875922 | 0.17 |
Gm45555 |
predicted gene 45555 |
2128 |
0.26 |
| chr10_127517746_127518141 | 0.17 |
Shmt2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
833 |
0.42 |
| chr6_24165121_24165289 | 0.17 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
2885 |
0.28 |
| chr11_16789776_16789927 | 0.17 |
Egfr |
epidermal growth factor receptor |
37621 |
0.14 |
| chr14_28656813_28657238 | 0.17 |
Gm35164 |
predicted gene, 35164 |
7266 |
0.27 |
| chr16_37630277_37630428 | 0.17 |
Hgd |
homogentisate 1, 2-dioxygenase |
8503 |
0.16 |
| chr7_25901958_25902263 | 0.16 |
Cyp2b10 |
cytochrome P450, family 2, subfamily b, polypeptide 10 |
4434 |
0.13 |
| chr18_38171641_38171818 | 0.16 |
Pcdh1 |
protocadherin 1 |
31434 |
0.11 |
| chr6_119354590_119354864 | 0.16 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
22165 |
0.17 |
| chr7_83634897_83635261 | 0.16 |
Gm45838 |
predicted gene 45838 |
1708 |
0.22 |
| chr7_66150558_66150709 | 0.16 |
Gm26827 |
predicted gene, 26827 |
22488 |
0.11 |
| chr6_108834552_108834703 | 0.16 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
1783 |
0.4 |
| chr12_25792666_25793052 | 0.16 |
Gm9321 |
predicted gene 9321 |
27682 |
0.22 |
| chr2_67902992_67903340 | 0.16 |
Gm37964 |
predicted gene, 37964 |
4570 |
0.31 |
| chr3_98742037_98742188 | 0.16 |
Hsd3b3 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
11353 |
0.13 |
| chr4_107422256_107422431 | 0.16 |
Gm24804 |
predicted gene, 24804 |
12229 |
0.15 |
| chr16_93856849_93857012 | 0.16 |
Morc3 |
microrchidia 3 |
3148 |
0.18 |
| chr8_70476439_70476616 | 0.16 |
Klhl26 |
kelch-like 26 |
378 |
0.72 |
| chr16_65977709_65977860 | 0.16 |
Gm49635 |
predicted gene, 49635 |
30678 |
0.21 |
| chr1_74018132_74018325 | 0.16 |
Tns1 |
tensin 1 |
1806 |
0.41 |
| chr11_120898760_120898937 | 0.16 |
Ccdc57 |
coiled-coil domain containing 57 |
12937 |
0.12 |
| chr2_79652088_79652307 | 0.16 |
Itprid2 |
ITPR interacting domain containing 2 |
13015 |
0.26 |
| chr7_31107633_31107788 | 0.15 |
Hpn |
hepsin |
3287 |
0.12 |
| chr4_115280301_115280685 | 0.15 |
Cyp4a12a |
cytochrome P450, family 4, subfamily a, polypeptide 12a |
18553 |
0.14 |
| chr11_11930894_11931056 | 0.15 |
Grb10 |
growth factor receptor bound protein 10 |
7641 |
0.19 |
| chr18_39710290_39710441 | 0.15 |
Gm50397 |
predicted gene, 50397 |
24594 |
0.19 |
| chr17_28330915_28331706 | 0.15 |
Rpl10a |
ribosomal protein L10A |
2032 |
0.16 |
| chr15_102456030_102456214 | 0.15 |
Gm24128 |
predicted gene, 24128 |
441 |
0.66 |
| chr13_54081500_54081675 | 0.15 |
Sfxn1 |
sideroflexin 1 |
4359 |
0.22 |
| chr8_109987720_109988100 | 0.15 |
Tat |
tyrosine aminotransferase |
2527 |
0.18 |
| chr15_55152954_55153171 | 0.15 |
Deptor |
DEP domain containing MTOR-interacting protein |
19609 |
0.17 |
| chr2_35052061_35052276 | 0.15 |
Hc |
hemolytic complement |
9270 |
0.16 |
| chr5_30248189_30248430 | 0.15 |
Gm16288 |
predicted gene 16288 |
7861 |
0.14 |
| chr7_97421325_97421669 | 0.15 |
Thrsp |
thyroid hormone responsive |
3767 |
0.16 |
| chr9_45204142_45204293 | 0.15 |
Tmprss4 |
transmembrane protease, serine 4 |
125 |
0.93 |
| chr1_74245666_74245817 | 0.15 |
Arpc2 |
actin related protein 2/3 complex, subunit 2 |
9191 |
0.09 |
| chr11_120980505_120980688 | 0.15 |
Csnk1d |
casein kinase 1, delta |
8971 |
0.1 |
| chr1_91456811_91456962 | 0.15 |
Per2 |
period circadian clock 2 |
2438 |
0.18 |
| chr2_178058332_178058492 | 0.14 |
Zfp931 |
zinc finger protein 931 |
20013 |
0.17 |
| chr11_60201956_60202762 | 0.14 |
Mir6922 |
microRNA 6922 |
32 |
0.63 |
| chr19_10066593_10066744 | 0.14 |
Fads2 |
fatty acid desaturase 2 |
4519 |
0.15 |
| chr14_7838492_7838643 | 0.14 |
Flnb |
filamin, beta |
20610 |
0.15 |
| chr16_57130407_57130558 | 0.14 |
Tomm70a |
translocase of outer mitochondrial membrane 70A |
7016 |
0.17 |
| chr17_66858572_66858789 | 0.14 |
Gm49940 |
predicted gene, 49940 |
3560 |
0.2 |
| chr12_8510428_8510853 | 0.14 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
561 |
0.71 |
| chr3_157897762_157897957 | 0.14 |
Cth |
cystathionase (cystathionine gamma-lyase) |
147 |
0.86 |
| chr2_152338144_152338307 | 0.14 |
Rbck1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
5572 |
0.1 |
| chr13_7307970_7308145 | 0.14 |
Gm8725 |
predicted gene 8725 |
255 |
0.95 |
| chr2_51142398_51142734 | 0.14 |
Rnd3 |
Rho family GTPase 3 |
6528 |
0.27 |
| chr2_122260835_122261566 | 0.14 |
Duox2 |
dual oxidase 2 |
24369 |
0.09 |
| chr19_16280365_16280542 | 0.14 |
Gm17068 |
predicted gene 17068 |
2656 |
0.25 |
| chr5_149635750_149636168 | 0.14 |
Hsph1 |
heat shock 105kDa/110kDa protein 1 |
212 |
0.91 |
| chr4_134511752_134511903 | 0.14 |
Aunip |
aurora kinase A and ninein interacting protein |
828 |
0.46 |
| chr4_149398701_149398906 | 0.14 |
Ube4b |
ubiquitination factor E4B |
575 |
0.68 |
| chr14_21422249_21422409 | 0.13 |
Gm25864 |
predicted gene, 25864 |
28145 |
0.17 |
| chr14_64154802_64154953 | 0.13 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
38563 |
0.11 |
| chr8_76535740_76535905 | 0.13 |
Gm27355 |
predicted gene, 27355 |
83460 |
0.1 |
| chr4_108070573_108070724 | 0.13 |
Scp2 |
sterol carrier protein 2, liver |
715 |
0.59 |
| chr5_28076176_28076405 | 0.13 |
Insig1 |
insulin induced gene 1 |
1624 |
0.34 |
| chr17_84650692_84651003 | 0.13 |
Dync2li1 |
dynein cytoplasmic 2 light intermediate chain 1 |
1572 |
0.29 |
| chr8_93098609_93098815 | 0.13 |
Ces1b |
carboxylesterase 1B |
18695 |
0.13 |
| chr10_115314574_115315134 | 0.13 |
Rab21 |
RAB21, member RAS oncogene family |
737 |
0.41 |
| chr1_73966648_73966904 | 0.13 |
Tns1 |
tensin 1 |
3733 |
0.28 |
| chr8_127135808_127135968 | 0.13 |
Pard3 |
par-3 family cell polarity regulator |
35516 |
0.19 |
| chr4_53105561_53105899 | 0.13 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
54165 |
0.12 |
| chr1_65155156_65155326 | 0.13 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
20323 |
0.11 |
| chr1_84237420_84237586 | 0.13 |
Mir6353 |
microRNA 6353 |
18542 |
0.21 |
| chr6_119373845_119373996 | 0.13 |
Adipor2 |
adiponectin receptor 2 |
14766 |
0.19 |
| chr8_70351602_70351757 | 0.13 |
Upf1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
1532 |
0.24 |
| chr12_10537211_10537367 | 0.13 |
Gm38407 |
predicted gene, 38407 |
10514 |
0.24 |
| chr13_54594427_54594595 | 0.13 |
Higd2a |
HIG1 domain family, member 2A |
4304 |
0.11 |
| chr4_148614904_148615132 | 0.13 |
Tardbp |
TAR DNA binding protein |
1120 |
0.34 |
| chr9_106247230_106247698 | 0.12 |
Alas1 |
aminolevulinic acid synthase 1 |
266 |
0.66 |
| chr8_89003653_89003846 | 0.12 |
Mir8110 |
microRNA 8110 |
20986 |
0.21 |
| chr14_120405158_120405340 | 0.12 |
Mbnl2 |
muscleblind like splicing factor 2 |
8762 |
0.22 |
| chrX_10719688_10719839 | 0.12 |
Gm14493 |
predicted gene 14493 |
1053 |
0.45 |
| chr2_58785851_58786309 | 0.12 |
Upp2 |
uridine phosphorylase 2 |
20755 |
0.19 |
| chr3_127103964_127104222 | 0.12 |
Ank2 |
ankyrin 2, brain |
20769 |
0.14 |
| chr4_106439943_106440414 | 0.12 |
Usp24 |
ubiquitin specific peptidase 24 |
11685 |
0.15 |
| chr16_16897078_16897229 | 0.12 |
Ppm1f |
protein phosphatase 1F (PP2C domain containing) |
579 |
0.57 |
| chr2_27757383_27757534 | 0.12 |
Rxra |
retinoid X receptor alpha |
17257 |
0.24 |
| chr13_34725685_34725960 | 0.12 |
Gm47151 |
predicted gene, 47151 |
5071 |
0.13 |
| chr2_109918614_109919408 | 0.12 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
1364 |
0.42 |
| chr5_143409630_143409781 | 0.12 |
Kdelr2 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
5867 |
0.09 |
| chr2_143923592_143923772 | 0.12 |
Dstn |
destrin |
8362 |
0.16 |
| chr2_168721341_168721659 | 0.12 |
Atp9a |
ATPase, class II, type 9A |
8538 |
0.2 |
| chr16_24642651_24642859 | 0.11 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
79100 |
0.1 |
| chr5_137376109_137376501 | 0.11 |
Zan |
zonadhesin |
25433 |
0.08 |
| chr15_55567278_55567443 | 0.11 |
Mtbp |
Mdm2, transformed 3T3 cell double minute p53 binding protein |
9420 |
0.2 |
| chr14_67234205_67234356 | 0.11 |
Ebf2 |
early B cell factor 2 |
364 |
0.78 |
| chr1_149758588_149758746 | 0.11 |
n-R5s218 |
nuclear encoded rRNA 5S 218 |
23809 |
0.2 |
| chr16_15637971_15638319 | 0.11 |
Prkdc |
protein kinase, DNA activated, catalytic polypeptide |
256 |
0.8 |
| chr9_9233329_9233549 | 0.11 |
Gm16833 |
predicted gene, 16833 |
2849 |
0.3 |
| chr18_21146229_21146380 | 0.11 |
Gm6378 |
predicted pseudogene 6378 |
69195 |
0.1 |
| chr6_6149620_6149797 | 0.11 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
24243 |
0.23 |
| chr12_103656604_103657009 | 0.11 |
Serpina6 |
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
296 |
0.83 |
| chr11_55204467_55204772 | 0.11 |
Slc36a1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
243 |
0.9 |
| chr7_100203719_100203885 | 0.11 |
Gm7067 |
predicted gene 7067 |
5454 |
0.14 |
| chr12_119426930_119427084 | 0.11 |
Macc1 |
metastasis associated in colon cancer 1 |
16403 |
0.25 |
| chr10_57488437_57488620 | 0.11 |
Hsf2 |
heat shock factor 2 |
2103 |
0.26 |
| chr2_29624791_29625100 | 0.11 |
Rapgef1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
5035 |
0.23 |
| chr6_54680979_54681130 | 0.11 |
Mturn |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
570 |
0.56 |
| chr11_120805486_120805657 | 0.11 |
Fasn |
fatty acid synthase |
3034 |
0.12 |
| chr1_72872241_72872412 | 0.11 |
Igfbp5 |
insulin-like growth factor binding protein 5 |
1949 |
0.37 |
| chr18_37806494_37806663 | 0.11 |
Pcdhgc3 |
protocadherin gamma subfamily C, 3 |
162 |
0.83 |
| chr1_131938675_131938875 | 0.11 |
Gm37676 |
predicted gene, 37676 |
5805 |
0.12 |
| chr7_29179934_29180147 | 0.11 |
Psmd8 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
441 |
0.61 |
| chr3_81966991_81967142 | 0.11 |
Tdo2 |
tryptophan 2,3-dioxygenase |
2983 |
0.21 |
| chr14_76901248_76901599 | 0.11 |
Gm48969 |
predicted gene, 48969 |
39852 |
0.15 |
| chr2_61492180_61492332 | 0.11 |
Gm22338 |
predicted gene, 22338 |
510 |
0.87 |
| chr11_74652093_74652447 | 0.10 |
Cluh |
clustered mitochondria (cluA/CLU1) homolog |
2440 |
0.25 |
| chr3_152248223_152248384 | 0.10 |
Nexn |
nexilin |
39 |
0.96 |
| chr10_93454032_93454309 | 0.10 |
Lta4h |
leukotriene A4 hydrolase |
719 |
0.62 |
| chr2_177501179_177501579 | 0.10 |
Gm14403 |
predicted gene 14403 |
3064 |
0.22 |
| chr1_136874160_136874323 | 0.10 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
66342 |
0.09 |
| chr9_23120915_23121075 | 0.10 |
Bmper |
BMP-binding endothelial regulator |
102081 |
0.08 |
| chr18_61827715_61827891 | 0.10 |
Afap1l1 |
actin filament associated protein 1-like 1 |
41101 |
0.13 |
| chr9_43219404_43219664 | 0.10 |
Rpl26-ps6 |
ribosomal protein L26, pseudogene 6 |
2412 |
0.22 |
| chr5_125493164_125493370 | 0.10 |
Aacs |
acetoacetyl-CoA synthetase |
12882 |
0.13 |
| chr13_35167228_35167404 | 0.10 |
4930529N20Rik |
RIKEN cDNA 4930529N20 gene |
488 |
0.52 |
| chr8_78656780_78656989 | 0.10 |
Slc10a7 |
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
40290 |
0.15 |
| chr16_37653013_37653243 | 0.10 |
Ndufb4 |
NADH:ubiquinone oxidoreductase subunit B4 |
1238 |
0.44 |
| chr2_27692193_27692459 | 0.10 |
Rxra |
retinoid X receptor alpha |
15064 |
0.24 |
| chr2_178098120_178098285 | 0.10 |
Zfp931 |
zinc finger protein 931 |
19726 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |