Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
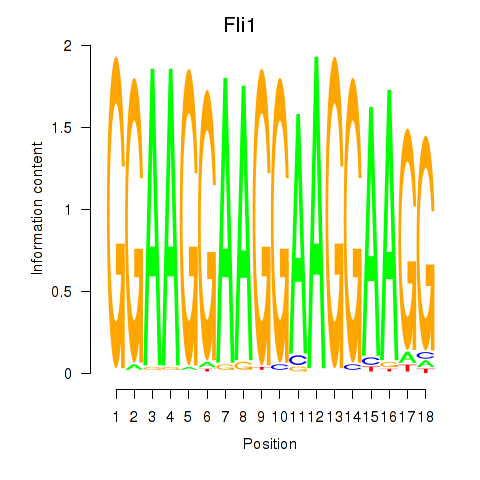
Results for Fli1
Z-value: 0.98
Transcription factors associated with Fli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fli1
|
ENSMUSG00000016087.7 | Friend leukemia integration 1 |
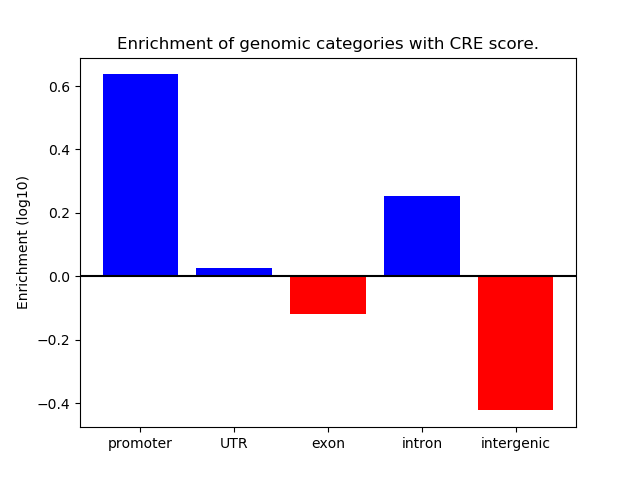
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_32541639_32541830 | Fli1 | 139 | 0.934809 | 0.71 | 1.2e-01 | Click! |
| chr9_32541322_32541500 | Fli1 | 184 | 0.913856 | 0.48 | 3.3e-01 | Click! |
| chr9_32540170_32540338 | Fli1 | 1341 | 0.305713 | 0.41 | 4.2e-01 | Click! |
| chr9_32544785_32544960 | Fli1 | 2011 | 0.224146 | 0.20 | 7.1e-01 | Click! |
Activity of the Fli1 motif across conditions
Conditions sorted by the z-value of the Fli1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
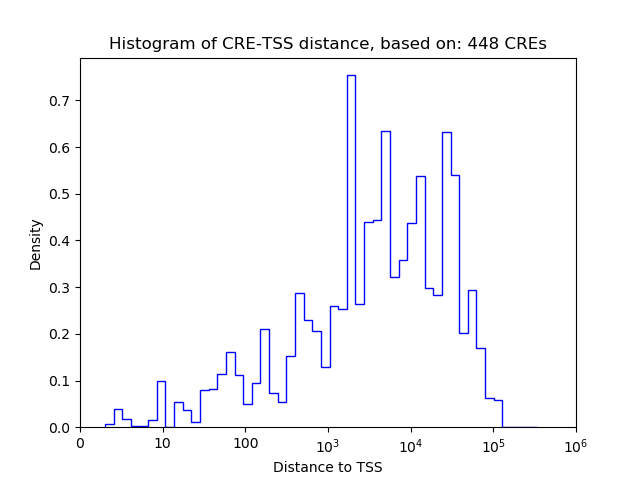
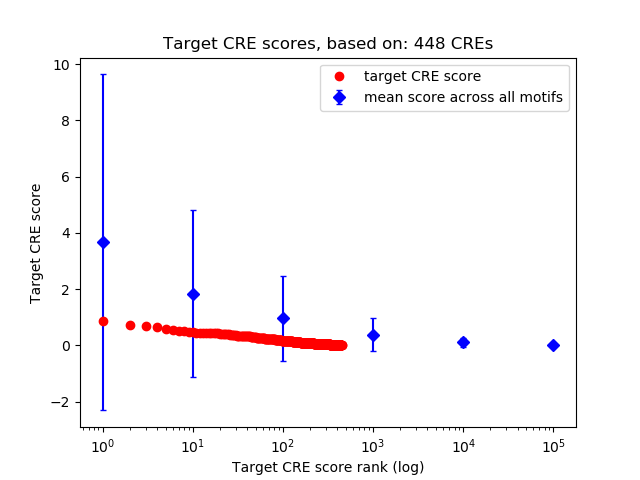
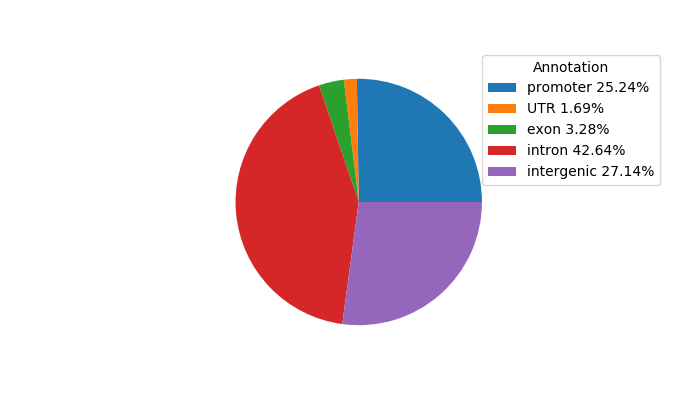
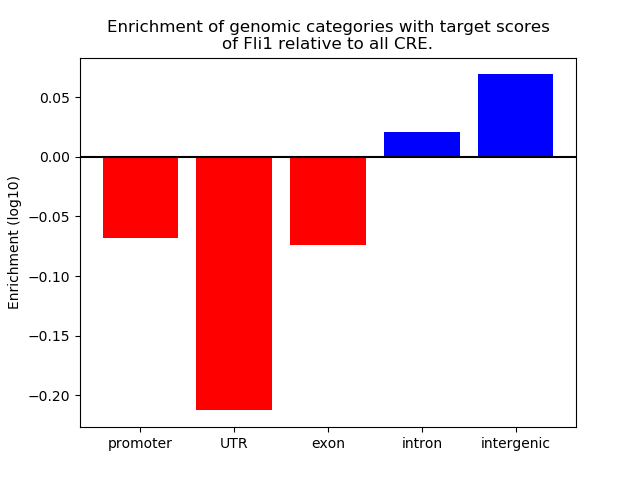
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_106528042_106528212 | 0.88 |
Gm22711 |
predicted gene, 22711 |
26882 |
0.13 |
| chr8_111739071_111739223 | 0.73 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
4662 |
0.21 |
| chr1_157417899_157418197 | 0.70 |
2810025M15Rik |
RIKEN cDNA 2810025M15 gene |
1854 |
0.24 |
| chr2_154615306_154615471 | 0.66 |
4930519P11Rik |
RIKEN cDNA 4930519P11 gene |
1951 |
0.18 |
| chr10_80539568_80539827 | 0.57 |
Atp8b3 |
ATPase, class I, type 8B, member 3 |
573 |
0.55 |
| chr8_110672217_110672368 | 0.53 |
Vac14 |
Vac14 homolog (S. cerevisiae) |
26712 |
0.15 |
| chr2_167254298_167254476 | 0.50 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
13783 |
0.14 |
| chr14_31957201_31957408 | 0.49 |
D830044D21Rik |
RIKEN cDNA D830044D21 gene |
50776 |
0.1 |
| chr9_75039237_75039563 | 0.48 |
Arpp19 |
cAMP-regulated phosphoprotein 19 |
1483 |
0.36 |
| chr9_32761430_32761581 | 0.47 |
Gm27240 |
predicted gene 27240 |
50334 |
0.11 |
| chr6_115996644_115996808 | 0.45 |
Plxnd1 |
plexin D1 |
1721 |
0.31 |
| chr5_129701203_129701458 | 0.45 |
Septin14 |
septin 14 |
2929 |
0.16 |
| chr18_75483489_75483815 | 0.44 |
Gm10532 |
predicted gene 10532 |
30993 |
0.21 |
| chr11_98770698_98771034 | 0.43 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
440 |
0.69 |
| chr2_67711621_67711818 | 0.43 |
Gm13604 |
predicted gene 13604 |
3948 |
0.2 |
| chr5_99669989_99670358 | 0.43 |
Gm16226 |
predicted gene 16226 |
12237 |
0.14 |
| chr11_68898151_68898311 | 0.43 |
Rpl26 |
ribosomal protein L26 |
3352 |
0.13 |
| chr7_18927869_18928042 | 0.43 |
Nova2 |
NOVA alternative splicing regulator 2 |
2067 |
0.18 |
| chr2_4510286_4510488 | 0.42 |
Frmd4a |
FERM domain containing 4A |
37193 |
0.15 |
| chr10_120895362_120895561 | 0.42 |
Msrb3 |
methionine sulfoxide reductase B3 |
3640 |
0.17 |
| chr11_63377101_63377266 | 0.41 |
B130011K05Rik |
RIKEN cDNA B130011K05 gene |
35986 |
0.18 |
| chr6_31544399_31544550 | 0.41 |
Podxl |
podocalyxin-like |
19507 |
0.17 |
| chr5_107046594_107046884 | 0.40 |
Gm33474 |
predicted gene, 33474 |
2097 |
0.34 |
| chr18_34989351_34989609 | 0.40 |
Gm36037 |
predicted gene, 36037 |
6510 |
0.15 |
| chr5_143678512_143678672 | 0.40 |
Cyth3 |
cytohesin 3 |
27351 |
0.14 |
| chr17_24645080_24645260 | 0.37 |
Slc9a3r2 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
180 |
0.85 |
| chr2_102968533_102968684 | 0.36 |
Cd44 |
CD44 antigen |
66943 |
0.1 |
| chr6_122826334_122826673 | 0.35 |
Foxj2 |
forkhead box J2 |
172 |
0.9 |
| chr11_19016500_19016765 | 0.35 |
Meis1 |
Meis homeobox 1 |
343 |
0.83 |
| chr8_105771287_105771438 | 0.35 |
Gfod2 |
glucose-fructose oxidoreductase domain containing 2 |
12698 |
0.1 |
| chr13_100543031_100543362 | 0.35 |
Ocln |
occludin |
9264 |
0.12 |
| chr1_178936614_178936788 | 0.34 |
Gm16564 |
predicted gene 16564 |
19087 |
0.24 |
| chr10_69925346_69925515 | 0.34 |
Ank3 |
ankyrin 3, epithelial |
65 |
0.99 |
| chr8_84189691_84189963 | 0.34 |
Nanos3 |
nanos C2HC-type zinc finger 3 |
4610 |
0.07 |
| chr18_61953404_61953581 | 0.34 |
Sh3tc2 |
SH3 domain and tetratricopeptide repeats 2 |
408 |
0.85 |
| chr13_44067362_44067519 | 0.33 |
Gm33630 |
predicted gene, 33630 |
32520 |
0.15 |
| chr13_37510108_37510259 | 0.33 |
Gm29590 |
predicted gene 29590 |
465 |
0.67 |
| chr16_94580932_94581197 | 0.32 |
Dyrk1a |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
10157 |
0.19 |
| chr17_26595433_26595607 | 0.32 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
644 |
0.59 |
| chr17_84037937_84038214 | 0.32 |
Gm49967 |
predicted gene, 49967 |
1185 |
0.36 |
| chr9_122084266_122084690 | 0.32 |
Snrk |
SNF related kinase |
32788 |
0.08 |
| chr2_92354348_92354505 | 0.31 |
Phf21a |
PHD finger protein 21A |
5515 |
0.12 |
| chr7_122404778_122404929 | 0.31 |
Gm44663 |
predicted gene 44663 |
2398 |
0.23 |
| chr17_4199793_4199983 | 0.31 |
4930548J01Rik |
RIKEN cDNA 4930548J01 gene |
77775 |
0.11 |
| chr1_138967552_138967703 | 0.31 |
Dennd1b |
DENN/MADD domain containing 1B |
93 |
0.91 |
| chr11_100392066_100392243 | 0.28 |
Jup |
junction plakoglobin |
5384 |
0.08 |
| chr3_122423925_122424117 | 0.28 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
4198 |
0.18 |
| chr7_84335830_84336210 | 0.28 |
Gm30873 |
predicted gene, 30873 |
2075 |
0.28 |
| chr1_107645210_107645382 | 0.28 |
Serpinb8 |
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
39416 |
0.15 |
| chr3_148907132_148907314 | 0.28 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
47412 |
0.16 |
| chr6_83475613_83475764 | 0.28 |
Dguok |
deoxyguanosine kinase |
11322 |
0.12 |
| chr6_53070500_53070680 | 0.27 |
Jazf1 |
JAZF zinc finger 1 |
1959 |
0.36 |
| chr2_131134497_131134751 | 0.27 |
Gm11037 |
predicted gene 11037 |
1127 |
0.32 |
| chr16_20371861_20372035 | 0.27 |
Abcc5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
4694 |
0.18 |
| chr11_16751759_16751915 | 0.26 |
Egfr |
epidermal growth factor receptor |
366 |
0.88 |
| chr2_72183135_72183304 | 0.26 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
3526 |
0.24 |
| chr14_7819506_7819687 | 0.26 |
Flnb |
filamin, beta |
1639 |
0.32 |
| chr9_70244291_70244584 | 0.25 |
Myo1e |
myosin IE |
37069 |
0.16 |
| chr2_93223637_93223793 | 0.25 |
Tspan18 |
tetraspanin 18 |
4071 |
0.26 |
| chr4_106575149_106575301 | 0.25 |
Gm12744 |
predicted gene 12744 |
13883 |
0.1 |
| chr3_132479376_132479701 | 0.25 |
Gm42880 |
predicted gene 42880 |
33990 |
0.19 |
| chr16_95964505_95964671 | 0.24 |
Psmg1 |
proteasome (prosome, macropain) assembly chaperone 1 |
25310 |
0.13 |
| chr10_68842447_68842629 | 0.24 |
1700048P04Rik |
RIKEN cDNA 1700048P04 gene |
61962 |
0.13 |
| chr3_61185919_61186073 | 0.24 |
Gm37719 |
predicted gene, 37719 |
105450 |
0.07 |
| chr4_43406064_43406489 | 0.24 |
Rusc2 |
RUN and SH3 domain containing 2 |
159 |
0.93 |
| chr12_80226238_80226389 | 0.24 |
Gm47767 |
predicted gene, 47767 |
17070 |
0.14 |
| chr6_87389476_87389814 | 0.23 |
Gkn3 |
gastrokine 3 |
710 |
0.57 |
| chr10_118797429_118797609 | 0.23 |
Gm4065 |
predicted gene 4065 |
7740 |
0.17 |
| chr10_86689713_86689885 | 0.23 |
Gm15344 |
predicted gene 15344 |
3296 |
0.09 |
| chr7_28442822_28442993 | 0.22 |
Gmfg |
glia maturation factor, gamma |
200 |
0.86 |
| chr3_59229138_59229406 | 0.22 |
P2ry12 |
purinergic receptor P2Y, G-protein coupled 12 |
1691 |
0.29 |
| chr17_15373087_15373287 | 0.22 |
Dll1 |
delta like canonical Notch ligand 1 |
885 |
0.54 |
| chr9_43220553_43220704 | 0.22 |
Oaf |
out at first homolog |
3346 |
0.18 |
| chr1_134238279_134238430 | 0.22 |
Adora1 |
adenosine A1 receptor |
2923 |
0.2 |
| chr2_144181951_144182219 | 0.21 |
Gm5535 |
predicted gene 5535 |
2999 |
0.21 |
| chr6_22004958_22005119 | 0.21 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
12028 |
0.25 |
| chr1_39735925_39736349 | 0.21 |
Rfx8 |
regulatory factor X 8 |
15140 |
0.19 |
| chr7_28440830_28441020 | 0.21 |
Gmfg |
glia maturation factor, gamma |
2 |
0.94 |
| chr1_125435331_125435561 | 0.21 |
Actr3 |
ARP3 actin-related protein 3 |
8 |
0.98 |
| chr9_21291584_21291735 | 0.21 |
Cdkn2d |
cyclin dependent kinase inhibitor 2D |
252 |
0.82 |
| chr4_13833049_13833208 | 0.21 |
Runx1t1 |
RUNX1 translocation partner 1 |
48346 |
0.18 |
| chr8_126593928_126594215 | 0.21 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
85 |
0.98 |
| chr4_136148351_136148658 | 0.20 |
Id3 |
inhibitor of DNA binding 3 |
3530 |
0.17 |
| chr10_81327977_81328184 | 0.20 |
Tbxa2r |
thromboxane A2 receptor |
651 |
0.39 |
| chr14_63247586_63247967 | 0.19 |
Gata4 |
GATA binding protein 4 |
2505 |
0.26 |
| chr5_122209998_122210149 | 0.19 |
Hvcn1 |
hydrogen voltage-gated channel 1 |
40 |
0.96 |
| chr11_100388061_100388239 | 0.19 |
Jup |
junction plakoglobin |
1380 |
0.21 |
| chr2_26493309_26493736 | 0.18 |
Notch1 |
notch 1 |
10300 |
0.1 |
| chr4_142084102_142084861 | 0.18 |
Tmem51os1 |
Tmem51 opposite strand 1 |
141 |
0.54 |
| chr14_121435885_121436040 | 0.18 |
Gm33299 |
predicted gene, 33299 |
19800 |
0.17 |
| chr5_150611809_150611960 | 0.18 |
Gm43597 |
predicted gene 43597 |
4881 |
0.11 |
| chr1_180330369_180330586 | 0.18 |
Itpkb |
inositol 1,4,5-trisphosphate 3-kinase B |
8 |
0.54 |
| chr17_15135586_15135747 | 0.18 |
Gm35455 |
predicted gene, 35455 |
16493 |
0.18 |
| chr4_47251829_47251990 | 0.18 |
Col15a1 |
collagen, type XV, alpha 1 |
6626 |
0.22 |
| chr4_122945793_122945944 | 0.18 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
14122 |
0.13 |
| chr18_64369558_64369811 | 0.18 |
Onecut2 |
one cut domain, family member 2 |
29664 |
0.13 |
| chr5_134680232_134680567 | 0.17 |
Limk1 |
LIM-domain containing, protein kinase |
2151 |
0.21 |
| chr4_81275942_81276093 | 0.17 |
Mpdz |
multiple PDZ domain protein |
8682 |
0.29 |
| chr11_120346748_120346946 | 0.17 |
Gm23419 |
predicted gene, 23419 |
28 |
0.58 |
| chr9_20760817_20761153 | 0.17 |
Olfm2 |
olfactomedin 2 |
14636 |
0.13 |
| chr17_47299700_47300022 | 0.17 |
Trerf1 |
transcriptional regulating factor 1 |
5792 |
0.18 |
| chr11_70179506_70179670 | 0.17 |
Clec10a |
C-type lectin domain family 10, member A |
10103 |
0.08 |
| chr11_120815283_120815743 | 0.17 |
Fasn |
fatty acid synthase |
58 |
0.94 |
| chr17_47567364_47567544 | 0.16 |
Ccnd3 |
cyclin D3 |
25982 |
0.1 |
| chr11_98794764_98794936 | 0.16 |
Msl1 |
male specific lethal 1 |
666 |
0.52 |
| chr15_22046405_22046937 | 0.16 |
Gm49187 |
predicted gene, 49187 |
88239 |
0.1 |
| chr5_53948666_53948853 | 0.16 |
Gm43266 |
predicted gene 43266 |
30315 |
0.16 |
| chr4_140673630_140673798 | 0.16 |
Gm13025 |
predicted gene 13025 |
6039 |
0.14 |
| chrX_9128548_9128703 | 0.16 |
Fthl17-ps3 |
ferritin, heavy polypeptide-like 17, pseudogene 3 |
5158 |
0.13 |
| chr18_35942087_35942447 | 0.16 |
Gm23882 |
predicted gene, 23882 |
10557 |
0.12 |
| chr11_108752151_108752318 | 0.16 |
Cep112 |
centrosomal protein 112 |
29 |
0.98 |
| chr15_35887277_35887437 | 0.15 |
Vps13b |
vacuolar protein sorting 13B |
15635 |
0.16 |
| chr1_100706996_100707453 | 0.15 |
Gm29334 |
predicted gene 29334 |
53424 |
0.15 |
| chr5_148789218_148789510 | 0.15 |
4930505K14Rik |
RIKEN cDNA 4930505K14 gene |
11401 |
0.15 |
| chr12_3888007_3888205 | 0.15 |
Dnmt3a |
DNA methyltransferase 3A |
2657 |
0.24 |
| chr1_55226495_55226826 | 0.15 |
Rftn2 |
raftlin family member 2 |
122 |
0.77 |
| chr6_38941118_38941304 | 0.15 |
Tbxas1 |
thromboxane A synthase 1, platelet |
190 |
0.95 |
| chr15_67146103_67146362 | 0.15 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
17209 |
0.24 |
| chr5_116024413_116024572 | 0.15 |
Prkab1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
16 |
0.96 |
| chr10_99467611_99467809 | 0.15 |
Gm35035 |
predicted gene, 35035 |
7480 |
0.16 |
| chr13_73352022_73352179 | 0.15 |
Eprn |
ephemeron, early developmental lncRNA |
4716 |
0.18 |
| chr4_82507892_82508053 | 0.14 |
Gm11266 |
predicted gene 11266 |
44 |
0.97 |
| chr2_119304857_119305179 | 0.14 |
Vps18 |
VPS18 CORVET/HOPS core subunit |
16231 |
0.1 |
| chr18_69660434_69660585 | 0.14 |
Tcf4 |
transcription factor 4 |
3842 |
0.35 |
| chr11_93988808_93989028 | 0.14 |
Spag9 |
sperm associated antigen 9 |
7173 |
0.15 |
| chr2_12197838_12198004 | 0.14 |
Itga8 |
integrin alpha 8 |
4609 |
0.27 |
| chr3_51301711_51301885 | 0.14 |
Gm37399 |
predicted gene, 37399 |
1105 |
0.37 |
| chr4_49536807_49537181 | 0.13 |
Aldob |
aldolase B, fructose-bisphosphate |
1872 |
0.25 |
| chr8_26078485_26078636 | 0.13 |
Hook3 |
hook microtubule tethering protein 3 |
817 |
0.46 |
| chr9_30906121_30906748 | 0.13 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
3223 |
0.28 |
| chr19_24017522_24017784 | 0.13 |
Gm50305 |
predicted gene, 50305 |
12873 |
0.17 |
| chr15_77745405_77745688 | 0.13 |
Gm49410 |
predicted gene, 49410 |
4785 |
0.11 |
| chr13_90896944_90897260 | 0.13 |
Atp6ap1l |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
1722 |
0.38 |
| chr2_169724307_169724473 | 0.13 |
Tshz2 |
teashirt zinc finger family member 2 |
90714 |
0.08 |
| chr3_96545689_96545844 | 0.13 |
Gm15441 |
predicted gene 15441 |
12049 |
0.07 |
| chr2_128704813_128704964 | 0.12 |
Mertk |
MER proto-oncogene tyrosine kinase |
5932 |
0.17 |
| chr5_23890845_23891003 | 0.12 |
AC117663.1 |
solute carrier family 40 (iron-regulated transporter), member 1 (Slc40a1), pseudogene |
14481 |
0.11 |
| chr16_4512630_4512833 | 0.12 |
Srl |
sarcalumenin |
10332 |
0.16 |
| chr2_170290505_170290775 | 0.12 |
Gm14270 |
predicted gene 14270 |
5605 |
0.25 |
| chr9_106371683_106372113 | 0.12 |
Dusp7 |
dual specificity phosphatase 7 |
1154 |
0.36 |
| chr4_108076318_108076469 | 0.12 |
Scp2 |
sterol carrier protein 2, liver |
4974 |
0.15 |
| chr3_89397517_89397668 | 0.11 |
Gm15417 |
predicted gene 15417 |
1596 |
0.15 |
| chr11_16900094_16900245 | 0.11 |
Egfr |
epidermal growth factor receptor |
5016 |
0.22 |
| chr7_84604824_84604975 | 0.11 |
Fah |
fumarylacetoacetate hydrolase |
830 |
0.56 |
| chr11_119685579_119685767 | 0.11 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
29729 |
0.15 |
| chr4_108040471_108040896 | 0.11 |
Gm23354 |
predicted gene, 23354 |
1117 |
0.39 |
| chr7_98084571_98084774 | 0.11 |
Myo7a |
myosin VIIA |
47 |
0.98 |
| chr11_3535115_3535308 | 0.11 |
Gm11947 |
predicted gene 11947 |
973 |
0.34 |
| chr2_118763590_118763758 | 0.10 |
Ccdc9b |
coiled-coil domain containing 9B |
1013 |
0.39 |
| chr1_174921018_174921617 | 0.10 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
502 |
0.88 |
| chr11_5510779_5510930 | 0.10 |
Xbp1 |
X-box binding protein 1 |
9805 |
0.13 |
| chr15_6909753_6910113 | 0.10 |
Osmr |
oncostatin M receptor |
34964 |
0.21 |
| chr10_28106282_28106464 | 0.10 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
31363 |
0.19 |
| chr13_110967143_110967402 | 0.10 |
Gm48547 |
predicted gene, 48547 |
51549 |
0.12 |
| chr8_123705299_123705474 | 0.10 |
6030466F02Rik |
RIKEN cDNA 6030466F02 gene |
28572 |
0.05 |
| chr2_169996026_169996202 | 0.10 |
AY702102 |
cDNA sequence AY702102 |
32927 |
0.22 |
| chr6_115996180_115996399 | 0.10 |
Plxnd1 |
plexin D1 |
1284 |
0.4 |
| chr13_104617090_104617248 | 0.10 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
67762 |
0.13 |
| chr2_122256897_122257048 | 0.10 |
Sord |
sorbitol dehydrogenase |
22223 |
0.09 |
| chr2_144178364_144178517 | 0.10 |
Gm5535 |
predicted gene 5535 |
646 |
0.67 |
| chr4_148602442_148602607 | 0.10 |
Masp2 |
mannan-binding lectin serine peptidase 2 |
30 |
0.95 |
| chr16_31201119_31201450 | 0.10 |
Acap2 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
39 |
0.97 |
| chr19_34745406_34745557 | 0.10 |
Slc16a12 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
1808 |
0.24 |
| chr19_11517487_11517638 | 0.09 |
Gm19261 |
predicted gene, 19261 |
609 |
0.45 |
| chr6_82997803_82997973 | 0.09 |
Gm43981 |
predicted gene, 43981 |
6243 |
0.08 |
| chr7_27039864_27040232 | 0.09 |
Gm4290 |
predicted gene 4290 |
6846 |
0.12 |
| chr2_38287183_38287356 | 0.09 |
Dennd1a |
DENN/MADD domain containing 1A |
51 |
0.96 |
| chr16_11269926_11270104 | 0.09 |
Mir1945 |
microRNA 1945 |
15570 |
0.12 |
| chr6_37431530_37431865 | 0.09 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
10449 |
0.27 |
| chr18_68006220_68006482 | 0.09 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
73094 |
0.09 |
| chr12_15651775_15651933 | 0.09 |
Gm4804 |
predicted gene 4804 |
45442 |
0.16 |
| chr19_34841997_34842148 | 0.09 |
Mir107 |
microRNA 107 |
21299 |
0.13 |
| chr4_44065513_44065687 | 0.09 |
Gne |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
1320 |
0.35 |
| chr5_90366149_90366302 | 0.09 |
Ankrd17 |
ankyrin repeat domain 17 |
3 |
0.87 |
| chr11_49196058_49196253 | 0.09 |
Zfp62 |
zinc finger protein 62 |
7137 |
0.12 |
| chr1_131096930_131097150 | 0.09 |
Mapkapk2 |
MAP kinase-activated protein kinase 2 |
786 |
0.54 |
| chr18_64643739_64644174 | 0.09 |
Atp8b1 |
ATPase, class I, type 8B, member 1 |
16851 |
0.13 |
| chr13_97297675_97297839 | 0.09 |
C430039J16Rik |
RIKEN cDNA C430039J16 gene |
14574 |
0.18 |
| chr16_15887069_15887635 | 0.09 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
66 |
0.97 |
| chr10_86992593_86993005 | 0.09 |
Gm16269 |
predicted gene 16269 |
7498 |
0.16 |
| chr2_128122505_128122660 | 0.09 |
Bcl2l11 |
BCL2-like 11 (apoptosis facilitator) |
3456 |
0.27 |
| chr2_11331635_11331790 | 0.09 |
Gm37851 |
predicted gene, 37851 |
1113 |
0.35 |
| chr14_121073081_121073251 | 0.08 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
28913 |
0.23 |
| chr12_3874325_3874480 | 0.08 |
Dnmt3a |
DNA methyltransferase 3A |
8211 |
0.17 |
| chr2_28630005_28630405 | 0.08 |
Gfi1b |
growth factor independent 1B |
8223 |
0.11 |
| chrX_109095601_109095821 | 0.08 |
Sh3bgrl |
SH3-binding domain glutamic acid-rich protein like |
202 |
0.94 |
| chr9_40192823_40193002 | 0.08 |
Zfp202 |
zinc finger protein 202 |
583 |
0.65 |
| chr18_35296224_35296431 | 0.08 |
Sil1 |
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
3488 |
0.23 |
| chr4_123667418_123667597 | 0.08 |
Gm12925 |
predicted gene 12925 |
53 |
0.96 |
| chr15_36879731_36879883 | 0.08 |
Gm10384 |
predicted gene 10384 |
9 |
0.97 |
| chr12_110510275_110510450 | 0.08 |
Gm19605 |
predicted gene, 19605 |
24154 |
0.14 |
| chr4_102571913_102572894 | 0.08 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
2308 |
0.44 |
| chr1_86046974_86047152 | 0.08 |
2810459M11Rik |
RIKEN cDNA 2810459M11 gene |
1200 |
0.33 |
| chr4_133967208_133967543 | 0.08 |
Hmgn2 |
high mobility group nucleosomal binding domain 2 |
66 |
0.96 |
| chr2_58765113_58765779 | 0.08 |
Upp2 |
uridine phosphorylase 2 |
121 |
0.97 |
| chr7_30143897_30144083 | 0.08 |
Gm10988 |
predicted gene 10988 |
7851 |
0.08 |
| chr3_79519137_79519302 | 0.08 |
Fnip2 |
folliculin interacting protein 2 |
3967 |
0.2 |
| chr9_78230201_78230424 | 0.08 |
Gsta1 |
glutathione S-transferase, alpha 1 (Ya) |
344 |
0.77 |
| chr3_98853787_98853964 | 0.08 |
Hsd3b1 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
5919 |
0.11 |
| chr17_46028764_46028966 | 0.07 |
Vegfa |
vascular endothelial growth factor A |
2491 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |