Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
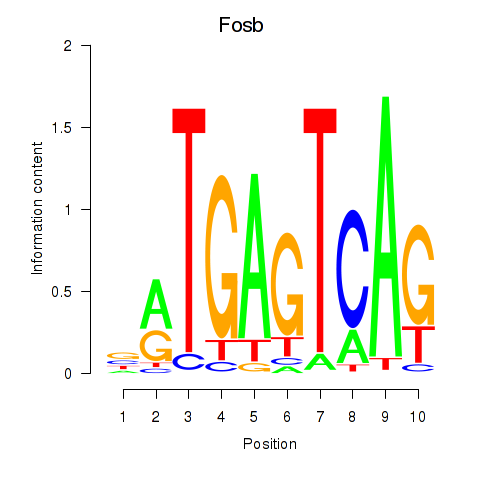
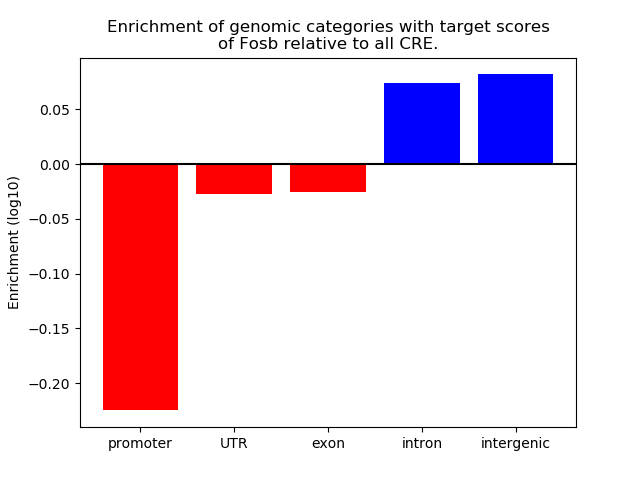
Results for Fosb
Z-value: 1.49
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSMUSG00000003545.2 | FBJ osteosarcoma oncogene B |
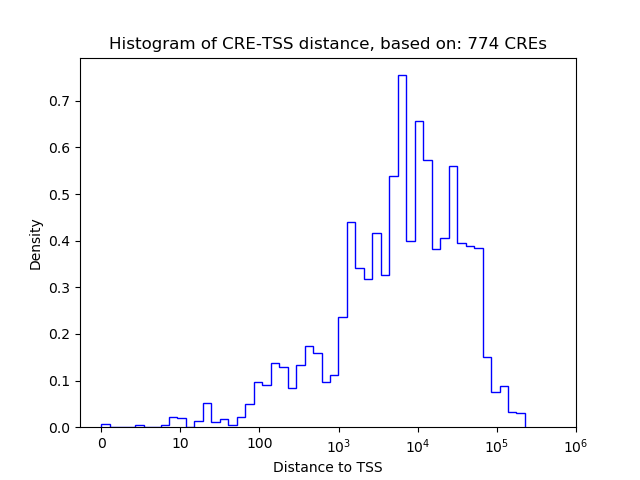
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_19320068_19320228 | Fosb | 10097 | 0.066494 | -0.85 | 3.4e-02 | Click! |
| chr7_19310289_19310447 | Fosb | 317 | 0.728755 | -0.75 | 8.5e-02 | Click! |
| chr7_19316542_19316693 | Fosb | 6566 | 0.071981 | -0.74 | 9.1e-02 | Click! |
| chr7_19308580_19308766 | Fosb | 927 | 0.303846 | -0.74 | 9.6e-02 | Click! |
| chr7_19309024_19309201 | Fosb | 488 | 0.566308 | -0.69 | 1.3e-01 | Click! |
Activity of the Fosb motif across conditions
Conditions sorted by the z-value of the Fosb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
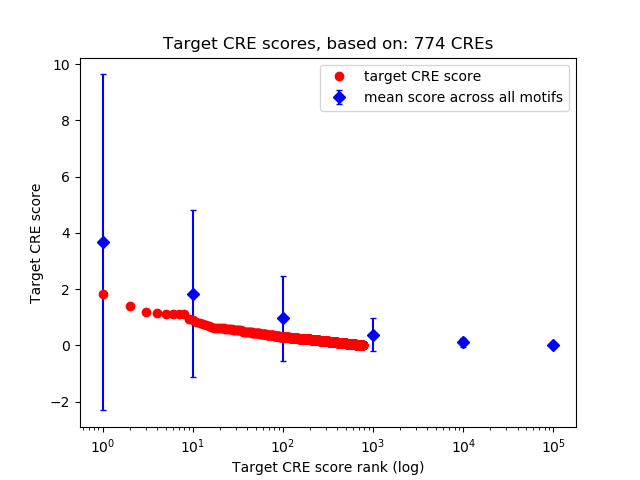
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_81319673_81319824 | 1.82 |
Gm30192 |
predicted gene, 30192 |
55040 |
0.14 |
| chr8_128648206_128648359 | 1.38 |
Gm26002 |
predicted gene, 26002 |
2715 |
0.24 |
| chr17_25127135_25127453 | 1.20 |
Clcn7 |
chloride channel, voltage-sensitive 7 |
6097 |
0.1 |
| chr8_128333459_128333621 | 1.15 |
Nrp1 |
neuropilin 1 |
25064 |
0.19 |
| chr11_16781411_16781603 | 1.12 |
Egfr |
epidermal growth factor receptor |
29277 |
0.16 |
| chr19_40153747_40153898 | 1.12 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr9_74947099_74947265 | 1.12 |
Fam214a |
family with sequence similarity 214, member A |
5702 |
0.22 |
| chr4_52815809_52815985 | 1.10 |
Olfr275 |
olfactory receptor 275 |
1293 |
0.37 |
| chr5_135693560_135693734 | 0.95 |
Por |
P450 (cytochrome) oxidoreductase |
4489 |
0.12 |
| chr15_96930772_96930923 | 0.88 |
Rpl10a-ps3 |
ribosomal protein L10A, pseudogene 3 |
7442 |
0.28 |
| chr17_64765537_64765696 | 0.84 |
Dreh |
down-regulated in hepatocellular carcinoma |
1495 |
0.41 |
| chr3_99188109_99188313 | 0.81 |
Gm18982 |
predicted gene, 18982 |
6930 |
0.13 |
| chrX_98051224_98051403 | 0.75 |
Gm14802 |
predicted gene 14802 |
10938 |
0.3 |
| chr9_44079482_44079643 | 0.73 |
Usp2 |
ubiquitin specific peptidase 2 |
5377 |
0.08 |
| chr1_178600841_178601326 | 0.70 |
Gm24405 |
predicted gene, 24405 |
35822 |
0.18 |
| chr4_6273243_6273469 | 0.65 |
Cyp7a1 |
cytochrome P450, family 7, subfamily a, polypeptide 1 |
2275 |
0.3 |
| chr15_3411405_3411556 | 0.63 |
Ghr |
growth hormone receptor |
60164 |
0.13 |
| chr10_87880054_87880205 | 0.62 |
Igf1os |
insulin-like growth factor 1, opposite strand |
16748 |
0.19 |
| chr5_135736099_135736335 | 0.62 |
Mir7034 |
microRNA 7034 |
328 |
0.49 |
| chr2_65267631_65267817 | 0.61 |
Gm13592 |
predicted gene 13592 |
9693 |
0.17 |
| chr10_111583961_111584156 | 0.61 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
10215 |
0.16 |
| chr13_31820560_31820717 | 0.60 |
Foxc1 |
forkhead box C1 |
14005 |
0.16 |
| chr8_126591993_126592171 | 0.59 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
1904 |
0.39 |
| chr2_130819037_130819547 | 0.59 |
4930402H24Rik |
RIKEN cDNA 4930402H24 gene |
5000 |
0.18 |
| chr18_20971049_20971351 | 0.58 |
Rnf125 |
ring finger protein 125 |
9728 |
0.2 |
| chr1_60833760_60833934 | 0.57 |
Gm11581 |
predicted gene 11581 |
24731 |
0.11 |
| chr13_113168591_113168750 | 0.56 |
Gzmk |
granzyme K |
12227 |
0.13 |
| chr14_47983137_47983357 | 0.56 |
Gm49304 |
predicted gene, 49304 |
10556 |
0.15 |
| chr3_18210346_18210497 | 0.56 |
Gm23686 |
predicted gene, 23686 |
32796 |
0.17 |
| chr12_104089620_104089844 | 0.55 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
9083 |
0.1 |
| chr1_162815471_162815637 | 0.54 |
Fmo4 |
flavin containing monooxygenase 4 |
1582 |
0.37 |
| chr16_70331018_70331193 | 0.54 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
16971 |
0.24 |
| chr12_104109060_104109241 | 0.54 |
Gm23289 |
predicted gene, 23289 |
133 |
0.92 |
| chr1_193870865_193871025 | 0.54 |
Gm21362 |
predicted gene, 21362 |
3936 |
0.37 |
| chr3_116784094_116784363 | 0.50 |
Agl |
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
2478 |
0.21 |
| chr12_105728790_105728966 | 0.49 |
Ak7 |
adenylate kinase 7 |
10022 |
0.17 |
| chr11_59226278_59226466 | 0.49 |
Arf1 |
ADP-ribosylation factor 1 |
1790 |
0.2 |
| chr3_61217484_61217722 | 0.48 |
Gm37719 |
predicted gene, 37719 |
73843 |
0.1 |
| chr11_4190659_4190839 | 0.48 |
Tbc1d10a |
TBC1 domain family, member 10a |
3927 |
0.12 |
| chr11_16836119_16836554 | 0.48 |
Egfros |
epidermal growth factor receptor, opposite strand |
5634 |
0.23 |
| chr9_96823694_96824186 | 0.48 |
Gm8520 |
predicted gene 8520 |
9606 |
0.15 |
| chr19_26753091_26753242 | 0.47 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
434 |
0.87 |
| chr2_167325906_167326068 | 0.46 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
12087 |
0.18 |
| chr15_3413229_3413380 | 0.46 |
Ghr |
growth hormone receptor |
58340 |
0.14 |
| chr2_148018834_148018985 | 0.46 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
19361 |
0.17 |
| chrX_60462043_60462214 | 0.45 |
Gm7073 |
predicted gene 7073 |
5933 |
0.23 |
| chr11_106490027_106490381 | 0.45 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
2352 |
0.23 |
| chr1_45672893_45673066 | 0.45 |
Gm23216 |
predicted gene, 23216 |
12583 |
0.19 |
| chr2_126867909_126868096 | 0.45 |
Gm14212 |
predicted gene 14212 |
3499 |
0.2 |
| chr15_3514160_3514323 | 0.44 |
Ghr |
growth hormone receptor |
42597 |
0.18 |
| chr3_88717858_88718018 | 0.43 |
Rit1 |
Ras-like without CAAX 1 |
1027 |
0.3 |
| chr9_116194113_116194264 | 0.43 |
Gm4668 |
predicted gene 4668 |
5008 |
0.18 |
| chr2_132656980_132657153 | 0.43 |
Rpl23a-ps4 |
ribosomal protein L23A, pseudogene 4 |
5176 |
0.13 |
| chr1_91463744_91463928 | 0.43 |
Per2 |
period circadian clock 2 |
4512 |
0.13 |
| chr12_79501822_79502159 | 0.42 |
Rad51b |
RAD51 paralog B |
174637 |
0.03 |
| chr3_137511395_137511566 | 0.42 |
Gm4861 |
predicted gene 4861 |
41142 |
0.16 |
| chr18_35813778_35813929 | 0.42 |
Ube2d2a |
ubiquitin-conjugating enzyme E2D 2A |
12546 |
0.09 |
| chr13_35434169_35434335 | 0.42 |
Gm48703 |
predicted gene, 48703 |
38869 |
0.17 |
| chr5_138788893_138789272 | 0.41 |
Fam20c |
family with sequence similarity 20, member C |
308 |
0.9 |
| chr10_121457902_121458053 | 0.40 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
4236 |
0.14 |
| chr7_98352590_98353259 | 0.40 |
Tsku |
tsukushi, small leucine rich proteoglycan |
7155 |
0.18 |
| chr16_93367939_93368144 | 0.40 |
1810044K17Rik |
RIKEN cDNA 1810044K17 gene |
198 |
0.89 |
| chr18_33335416_33335586 | 0.39 |
Gm5503 |
predicted gene 5503 |
49454 |
0.16 |
| chr1_70918439_70918607 | 0.39 |
Gm16236 |
predicted gene 16236 |
121507 |
0.06 |
| chr15_42674825_42675078 | 0.39 |
Gm17473 |
predicted gene, 17473 |
1290 |
0.44 |
| chr3_145995608_145995781 | 0.38 |
Syde2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
2023 |
0.3 |
| chr8_79710908_79711237 | 0.38 |
Abce1 |
ATP-binding cassette, sub-family E (OABP), member 1 |
578 |
0.48 |
| chr12_104850467_104850964 | 0.37 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
6656 |
0.19 |
| chr9_103507084_103507251 | 0.37 |
Tmem108 |
transmembrane protein 108 |
13556 |
0.1 |
| chr4_139601582_139602248 | 0.37 |
Gm21969 |
predicted gene 21969 |
1271 |
0.37 |
| chr1_85246945_85247096 | 0.36 |
Gm16028 |
predicted gene 16028 |
169 |
0.83 |
| chr1_137034426_137034730 | 0.36 |
Gm23763 |
predicted gene, 23763 |
44308 |
0.16 |
| chr17_10702814_10703005 | 0.36 |
Pacrg |
PARK2 co-regulated |
71842 |
0.11 |
| chr7_51868214_51868365 | 0.36 |
Gas2 |
growth arrest specific 2 |
4385 |
0.17 |
| chr14_89610572_89610930 | 0.36 |
Gm25415 |
predicted gene, 25415 |
96054 |
0.09 |
| chr18_8327970_8328137 | 0.35 |
Gm5500 |
predicted pseudogene 5500 |
87076 |
0.09 |
| chr15_58972603_58972890 | 0.35 |
Mtss1 |
MTSS I-BAR domain containing 1 |
207 |
0.92 |
| chr6_108313163_108313314 | 0.35 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
43016 |
0.18 |
| chr11_86629518_86629733 | 0.35 |
Gm11478 |
predicted gene 11478 |
1401 |
0.38 |
| chr2_142804438_142804612 | 0.35 |
Gm22310 |
predicted gene, 22310 |
17629 |
0.25 |
| chr4_108363811_108364169 | 0.34 |
Shisal2a |
shisa like 2A |
19359 |
0.12 |
| chr6_91712952_91713256 | 0.34 |
Gm44981 |
predicted gene 44981 |
174 |
0.92 |
| chr11_119777405_119777556 | 0.34 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
21293 |
0.16 |
| chr18_38406873_38407073 | 0.34 |
Ndfip1 |
Nedd4 family interacting protein 1 |
3423 |
0.18 |
| chr7_118104788_118104966 | 0.34 |
Rps15a |
ribosomal protein S15A |
10351 |
0.17 |
| chr1_67187795_67187968 | 0.33 |
Gm15668 |
predicted gene 15668 |
61319 |
0.11 |
| chr18_12686874_12687043 | 0.33 |
Ttc39c |
tetratricopeptide repeat domain 39C |
2770 |
0.21 |
| chr4_84131285_84131460 | 0.33 |
Gm12414 |
predicted gene 12414 |
1083 |
0.59 |
| chr18_53864446_53864629 | 0.32 |
Csnk1g3 |
casein kinase 1, gamma 3 |
2349 |
0.42 |
| chr1_10604540_10604691 | 0.32 |
Gm25253 |
predicted gene, 25253 |
48955 |
0.13 |
| chr15_58965948_58966356 | 0.32 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6387 |
0.16 |
| chr2_85061178_85061331 | 0.31 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
290 |
0.85 |
| chr2_59997998_59998149 | 0.31 |
Gm13574 |
predicted gene 13574 |
5729 |
0.19 |
| chr5_140616803_140616967 | 0.31 |
Lfng |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
5200 |
0.14 |
| chr14_20291279_20291481 | 0.31 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
3317 |
0.16 |
| chr6_71420889_71421080 | 0.31 |
Gm44130 |
predicted gene, 44130 |
14123 |
0.1 |
| chr18_85490238_85490398 | 0.31 |
Gm7612 |
predicted gene 7612 |
226407 |
0.02 |
| chr14_17900415_17900617 | 0.31 |
Thrb |
thyroid hormone receptor beta |
62305 |
0.13 |
| chr12_79864462_79864614 | 0.30 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
60195 |
0.12 |
| chr4_117937047_117937475 | 0.30 |
Artn |
artemin |
7498 |
0.12 |
| chr14_74650101_74650379 | 0.30 |
Htr2a |
5-hydroxytryptamine (serotonin) receptor 2A |
9400 |
0.24 |
| chr2_18048778_18049088 | 0.30 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr17_23554288_23554546 | 0.30 |
Casp16 |
caspase 16, apoptosis-related cysteine peptidase |
929 |
0.34 |
| chr2_74100092_74100243 | 0.30 |
A630050E04Rik |
RIKEN cDNA A630050E04 gene |
2175 |
0.32 |
| chr11_60240852_60241067 | 0.30 |
Gm12265 |
predicted gene 12265 |
149 |
0.92 |
| chr11_70972275_70972426 | 0.30 |
Rpain |
RPA interacting protein |
453 |
0.66 |
| chr19_14692311_14692469 | 0.30 |
Gm26026 |
predicted gene, 26026 |
26090 |
0.21 |
| chr19_27492565_27492719 | 0.30 |
Gm50101 |
predicted gene, 50101 |
19866 |
0.23 |
| chr8_80962240_80962391 | 0.29 |
Gm27048 |
predicted gene, 27048 |
23648 |
0.15 |
| chr14_31112566_31112718 | 0.29 |
Smim4 |
small integral membrane protein 4 |
16196 |
0.1 |
| chr6_136938692_136939081 | 0.29 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
705 |
0.59 |
| chr19_8757228_8757608 | 0.29 |
Nxf1 |
nuclear RNA export factor 1 |
268 |
0.68 |
| chr2_130488707_130488873 | 0.28 |
9530056E24Rik |
RIKEN cDNA 9530056E24 gene |
35205 |
0.08 |
| chr7_137621172_137621473 | 0.28 |
Gm45683 |
predicted gene 45683 |
27767 |
0.21 |
| chr4_76424963_76425155 | 0.28 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
24921 |
0.18 |
| chr19_39282868_39283045 | 0.28 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
4118 |
0.26 |
| chr2_168524880_168525160 | 0.28 |
Nfatc2 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
27256 |
0.2 |
| chr7_39977983_39978150 | 0.28 |
Gm44992 |
predicted gene 44992 |
8 |
0.98 |
| chr19_15007147_15007306 | 0.27 |
Gm5513 |
predicted pseudogene 5513 |
46460 |
0.21 |
| chr8_61585158_61585376 | 0.27 |
Palld |
palladin, cytoskeletal associated protein |
5872 |
0.3 |
| chr2_129582332_129582507 | 0.27 |
Sirpa |
signal-regulatory protein alpha |
10416 |
0.17 |
| chr8_90870510_90870662 | 0.27 |
Gm45640 |
predicted gene 45640 |
5995 |
0.14 |
| chr2_31742282_31742433 | 0.27 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
17586 |
0.14 |
| chr16_91433633_91433806 | 0.27 |
Gm46562 |
predicted gene, 46562 |
24702 |
0.08 |
| chrX_10259432_10259607 | 0.26 |
Otc |
ornithine transcarbamylase |
7152 |
0.26 |
| chr11_98294355_98294506 | 0.26 |
Gm20644 |
predicted gene 20644 |
9048 |
0.1 |
| chr4_105557523_105557702 | 0.26 |
Gm12726 |
predicted gene 12726 |
20728 |
0.24 |
| chr6_87277564_87277945 | 0.26 |
Gm44418 |
predicted gene, 44418 |
9 |
0.96 |
| chr16_38289983_38290198 | 0.26 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
4734 |
0.16 |
| chr19_21820366_21820517 | 0.26 |
Gm50130 |
predicted gene, 50130 |
13315 |
0.21 |
| chr7_99173461_99173666 | 0.26 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
3457 |
0.17 |
| chr5_66035556_66035796 | 0.25 |
Rbm47 |
RNA binding motif protein 47 |
18876 |
0.11 |
| chr9_25504836_25504987 | 0.25 |
Eepd1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
22044 |
0.17 |
| chr8_124014864_124015025 | 0.25 |
Taf5l |
TATA-box binding protein associated factor 5 like |
5996 |
0.11 |
| chr8_56230092_56230281 | 0.25 |
Hpgd |
hydroxyprostaglandin dehydrogenase 15 (NAD) |
64399 |
0.12 |
| chr11_16914824_16915305 | 0.25 |
Egfr |
epidermal growth factor receptor |
9879 |
0.18 |
| chr6_84043664_84043821 | 0.25 |
Gm7498 |
predicted gene 7498 |
12154 |
0.15 |
| chr7_18948266_18948455 | 0.25 |
Nova2 |
NOVA alternative splicing regulator 2 |
22472 |
0.08 |
| chr10_66928213_66928377 | 0.25 |
Gm26576 |
predicted gene, 26576 |
7993 |
0.17 |
| chr1_58729553_58729738 | 0.25 |
Cflar |
CASP8 and FADD-like apoptosis regulator |
907 |
0.49 |
| chr16_59956556_59956707 | 0.25 |
Epha6 |
Eph receptor A6 |
49090 |
0.18 |
| chr15_84924832_84925200 | 0.25 |
Nup50 |
nucleoporin 50 |
1567 |
0.26 |
| chr11_16702482_16702935 | 0.25 |
Gm25698 |
predicted gene, 25698 |
30003 |
0.15 |
| chr4_108234244_108234395 | 0.24 |
Zyg11b |
zyg-ll family member B, cell cycle regulator |
7719 |
0.16 |
| chr11_101464329_101464518 | 0.24 |
Rnd2 |
Rho family GTPase 2 |
576 |
0.46 |
| chr2_32517571_32517751 | 0.24 |
Gm13412 |
predicted gene 13412 |
7370 |
0.1 |
| chr16_30375250_30375569 | 0.24 |
Atp13a3 |
ATPase type 13A3 |
13076 |
0.2 |
| chr13_101540438_101540589 | 0.24 |
Gm47533 |
predicted gene, 47533 |
4693 |
0.19 |
| chr9_68009384_68009551 | 0.24 |
Gm20745 |
predicted gene, 20745 |
16247 |
0.23 |
| chr19_42017962_42018130 | 0.24 |
Ubtd1 |
ubiquitin domain containing 1 |
897 |
0.43 |
| chr19_12633216_12633544 | 0.24 |
Glyat |
glycine-N-acyltransferase |
72 |
0.94 |
| chr5_21702946_21703133 | 0.24 |
Napepld |
N-acyl phosphatidylethanolamine phospholipase D |
1643 |
0.28 |
| chr15_58947619_58947948 | 0.24 |
Mtss1 |
MTSS I-BAR domain containing 1 |
5797 |
0.14 |
| chr16_77535131_77535288 | 0.24 |
Gm36963 |
predicted gene, 36963 |
4673 |
0.14 |
| chr16_20540431_20540616 | 0.24 |
Ap2m1 |
adaptor-related protein complex 2, mu 1 subunit |
100 |
0.9 |
| chr3_149220840_149220991 | 0.24 |
Gm10287 |
predicted gene 10287 |
4830 |
0.24 |
| chr11_106410149_106410681 | 0.24 |
Icam2 |
intercellular adhesion molecule 2 |
22340 |
0.13 |
| chr16_55967011_55967191 | 0.24 |
Rpl24 |
ribosomal protein L24 |
554 |
0.46 |
| chr4_117126763_117126969 | 0.24 |
Tctex1d4 |
Tctex1 domain containing 4 |
53 |
0.9 |
| chr14_76715025_76715203 | 0.24 |
1700108F19Rik |
RIKEN cDNA 1700108F19 gene |
33360 |
0.15 |
| chr12_33369704_33369869 | 0.24 |
Atxn7l1 |
ataxin 7-like 1 |
2462 |
0.24 |
| chr5_88367022_88367405 | 0.24 |
Amtn |
amelotin |
8895 |
0.17 |
| chr1_35759768_35759937 | 0.24 |
Gm25634 |
predicted gene, 25634 |
1750 |
0.42 |
| chr4_140862219_140862586 | 0.24 |
4930515B02Rik |
RIKEN cDNA 4930515B02 gene |
10291 |
0.12 |
| chr14_89612108_89612276 | 0.23 |
Gm25415 |
predicted gene, 25415 |
94613 |
0.09 |
| chr19_40227668_40227819 | 0.23 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
3062 |
0.2 |
| chr1_4670727_4670906 | 0.23 |
Gm6085 |
predicted gene 6085 |
18587 |
0.13 |
| chr9_122856843_122857012 | 0.23 |
Zfp445 |
zinc finger protein 445 |
200 |
0.89 |
| chr6_121145100_121145251 | 0.23 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
14176 |
0.14 |
| chr15_59068526_59068830 | 0.23 |
Mtss1 |
MTSS I-BAR domain containing 1 |
13214 |
0.22 |
| chr4_106439943_106440414 | 0.23 |
Usp24 |
ubiquitin specific peptidase 24 |
11685 |
0.15 |
| chr3_27712516_27712706 | 0.23 |
Fndc3b |
fibronectin type III domain containing 3B |
1304 |
0.58 |
| chr15_62743599_62743898 | 0.23 |
Gm22521 |
predicted gene, 22521 |
49453 |
0.17 |
| chr10_80342748_80342921 | 0.23 |
Adamtsl5 |
ADAMTS-like 5 |
2404 |
0.1 |
| chr12_34664877_34665034 | 0.22 |
Gm47357 |
predicted gene, 47357 |
36317 |
0.19 |
| chr10_59884883_59885114 | 0.22 |
Dnajb12 |
DnaJ heat shock protein family (Hsp40) member B12 |
5372 |
0.16 |
| chr7_128529551_128529973 | 0.22 |
Bag3 |
BCL2-associated athanogene 3 |
6146 |
0.12 |
| chr14_47533422_47533895 | 0.22 |
Fbxo34 |
F-box protein 34 |
7579 |
0.12 |
| chr1_177188880_177189041 | 0.22 |
Gm37706 |
predicted gene, 37706 |
18098 |
0.16 |
| chr8_111246376_111246527 | 0.22 |
Glg1 |
golgi apparatus protein 1 |
12749 |
0.16 |
| chr11_12463629_12463800 | 0.22 |
Cobl |
cordon-bleu WH2 repeat |
1038 |
0.69 |
| chr9_65198022_65198189 | 0.22 |
Gm25313 |
predicted gene, 25313 |
1418 |
0.27 |
| chr8_64877797_64877973 | 0.22 |
Klhl2 |
kelch-like 2, Mayven |
27868 |
0.11 |
| chr15_62337746_62337903 | 0.22 |
Pvt1 |
Pvt1 oncogene |
115221 |
0.06 |
| chr2_27727019_27727170 | 0.22 |
Rxra |
retinoid X receptor alpha |
1875 |
0.44 |
| chr3_62458537_62458746 | 0.22 |
Dhx36 |
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
13946 |
0.24 |
| chr11_101111260_101111411 | 0.22 |
Retreg3 |
reticulophagy regulator family member 3 |
8430 |
0.08 |
| chr17_80772516_80772751 | 0.22 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
44148 |
0.15 |
| chr1_91445260_91445618 | 0.22 |
Per2 |
period circadian clock 2 |
4426 |
0.13 |
| chr11_120821685_120822756 | 0.22 |
Fasn |
fatty acid synthase |
1560 |
0.22 |
| chr10_81409520_81409671 | 0.22 |
Gm16104 |
predicted gene 16104 |
20 |
0.92 |
| chr8_72134846_72135406 | 0.22 |
Tpm4 |
tropomyosin 4 |
103 |
0.92 |
| chr4_108090689_108090857 | 0.21 |
Mir6397 |
microRNA 6397 |
3416 |
0.18 |
| chr16_43464306_43464503 | 0.21 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
39210 |
0.15 |
| chr2_105252035_105252217 | 0.21 |
Them7 |
thioesterase superfamily member 7 |
27784 |
0.2 |
| chr16_35484667_35484833 | 0.21 |
Pdia5 |
protein disulfide isomerase associated 5 |
5839 |
0.17 |
| chr8_107293609_107293943 | 0.21 |
Nfat5 |
nuclear factor of activated T cells 5 |
217 |
0.94 |
| chr12_75545967_75546292 | 0.21 |
Ppp2r5e |
protein phosphatase 2, regulatory subunit B', epsilon |
3545 |
0.25 |
| chr6_108510231_108510410 | 0.21 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
2007 |
0.27 |
| chr3_52296364_52296605 | 0.21 |
Gm38034 |
predicted gene, 38034 |
14243 |
0.14 |
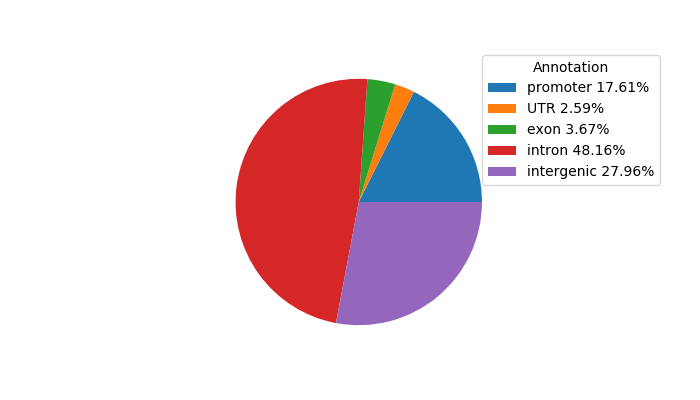
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.0 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | Genes involved in Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |