Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
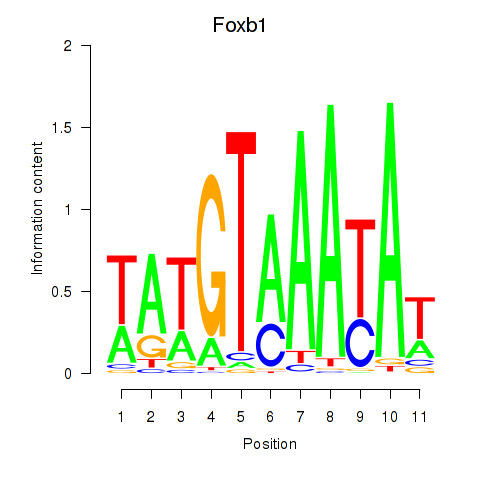
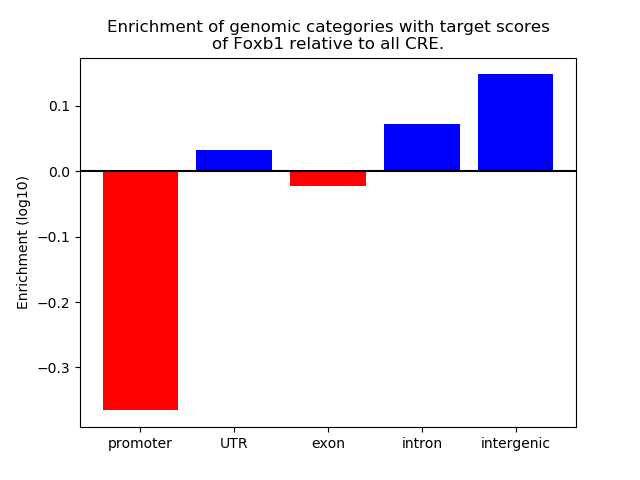
Results for Foxb1
Z-value: 1.39
Transcription factors associated with Foxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxb1
|
ENSMUSG00000059246.4 | forkhead box B1 |
Activity of the Foxb1 motif across conditions
Conditions sorted by the z-value of the Foxb1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
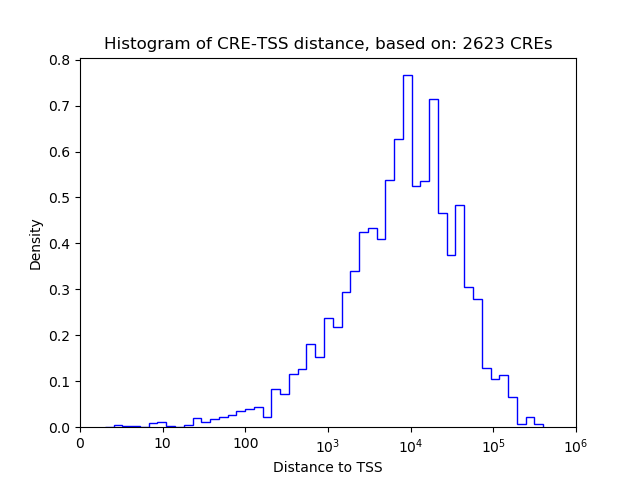
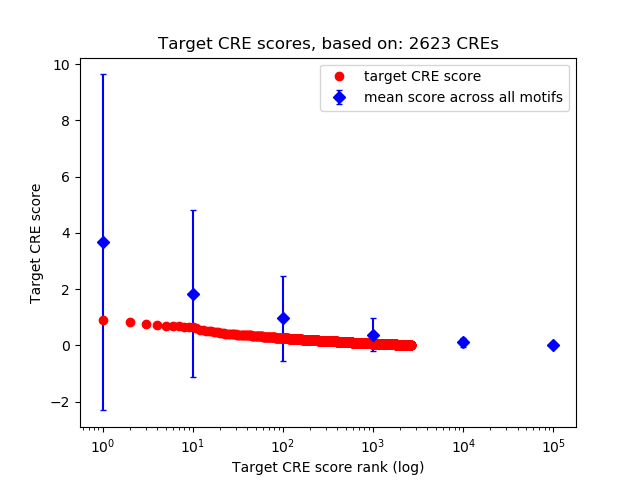
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_16888258_16888409 | 0.91 |
Egfr |
epidermal growth factor receptor |
10183 |
0.2 |
| chr2_8124301_8124468 | 0.84 |
Gm13254 |
predicted gene 13254 |
23481 |
0.29 |
| chr11_16588986_16589143 | 0.76 |
Gm12663 |
predicted gene 12663 |
47002 |
0.12 |
| chr7_26781568_26782082 | 0.71 |
Cyp2b19 |
cytochrome P450, family 2, subfamily b, polypeptide 19 |
15270 |
0.15 |
| chr2_25384633_25384784 | 0.69 |
Sapcd2 |
suppressor APC domain containing 2 |
8748 |
0.07 |
| chr16_45168608_45168759 | 0.68 |
Atg3 |
autophagy related 3 |
2800 |
0.24 |
| chr17_40806286_40806473 | 0.67 |
Crisp2 |
cysteine-rich secretory protein 2 |
624 |
0.67 |
| chrX_47259642_47259793 | 0.65 |
Gm14609 |
predicted gene 14609 |
121617 |
0.06 |
| chr4_65080374_65080537 | 0.65 |
Pappa |
pregnancy-associated plasma protein A |
43719 |
0.19 |
| chr13_56648096_56648259 | 0.64 |
Tgfbi |
transforming growth factor, beta induced |
16964 |
0.19 |
| chr7_99899121_99899281 | 0.60 |
Xrra1 |
X-ray radiation resistance associated 1 |
3505 |
0.15 |
| chr9_114632451_114632621 | 0.55 |
Cnot10 |
CCR4-NOT transcription complex, subunit 10 |
7427 |
0.16 |
| chr5_150264404_150264568 | 0.53 |
Fry |
FRY microtubule binding protein |
4719 |
0.21 |
| chr6_141381408_141381711 | 0.52 |
Gm10400 |
predicted gene 10400 |
41006 |
0.17 |
| chr19_20723027_20723214 | 0.51 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
4442 |
0.3 |
| chr10_71193153_71193343 | 0.51 |
1700113B09Rik |
RIKEN cDNA 1700113B09 gene |
8497 |
0.12 |
| chr5_148936597_148936854 | 0.47 |
Katnal1 |
katanin p60 subunit A-like 1 |
7405 |
0.1 |
| chr15_10972894_10973115 | 0.47 |
Amacr |
alpha-methylacyl-CoA racemase |
8752 |
0.15 |
| chr1_162986828_162987029 | 0.46 |
Fmo3 |
flavin containing monooxygenase 3 |
2400 |
0.25 |
| chr8_40907855_40908028 | 0.45 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
9503 |
0.16 |
| chr9_20745269_20745579 | 0.45 |
Olfm2 |
olfactomedin 2 |
925 |
0.51 |
| chr4_87880355_87880506 | 0.43 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
5888 |
0.31 |
| chr7_97423614_97423975 | 0.42 |
Thrsp |
thyroid hormone responsive |
6064 |
0.14 |
| chr8_73054856_73055038 | 0.41 |
Mir28b |
microRNA 28b |
38435 |
0.17 |
| chr7_84212215_84212389 | 0.41 |
Gm44826 |
predicted gene 44826 |
1788 |
0.29 |
| chrX_106036704_106036855 | 0.41 |
Atp7a |
ATPase, Cu++ transporting, alpha polypeptide |
9457 |
0.17 |
| chr8_114154739_114154891 | 0.41 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
21173 |
0.24 |
| chr19_10590708_10591077 | 0.40 |
Cyb561a3 |
cytochrome b561 family, member A3 |
12880 |
0.08 |
| chr11_107270789_107270983 | 0.40 |
Gm11719 |
predicted gene 11719 |
59822 |
0.08 |
| chr19_40146515_40146666 | 0.39 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
40696 |
0.11 |
| chr11_61274046_61274219 | 0.38 |
Aldh3a2 |
aldehyde dehydrogenase family 3, subfamily A2 |
6668 |
0.17 |
| chr16_4211280_4211431 | 0.38 |
Crebbp |
CREB binding protein |
2058 |
0.29 |
| chr15_53163055_53163223 | 0.38 |
Ext1 |
exostosin glycosyltransferase 1 |
33116 |
0.24 |
| chr5_123851097_123851267 | 0.38 |
Hcar2 |
hydroxycarboxylic acid receptor 2 |
14317 |
0.13 |
| chr13_66949557_66949708 | 0.37 |
Ptdss1 |
phosphatidylserine synthase 1 |
4309 |
0.12 |
| chr9_25795226_25795396 | 0.37 |
Gm48337 |
predicted gene, 48337 |
16436 |
0.17 |
| chr19_37077879_37078030 | 0.36 |
Gm22714 |
predicted gene, 22714 |
71208 |
0.08 |
| chr1_67233878_67234117 | 0.36 |
Gm15668 |
predicted gene 15668 |
15203 |
0.21 |
| chr17_4479666_4479822 | 0.36 |
4930517M08Rik |
RIKEN cDNA 4930517M08 gene |
155234 |
0.04 |
| chr11_16808080_16808304 | 0.36 |
Egfros |
epidermal growth factor receptor, opposite strand |
22510 |
0.19 |
| chr18_53860444_53860644 | 0.36 |
Csnk1g3 |
casein kinase 1, gamma 3 |
1578 |
0.52 |
| chr5_51039736_51040095 | 0.36 |
Gm40319 |
predicted gene, 40319 |
12466 |
0.29 |
| chr15_9602541_9602716 | 0.35 |
Il7r |
interleukin 7 receptor |
72452 |
0.11 |
| chr1_66839875_66840046 | 0.35 |
Gm15793 |
predicted gene 15793 |
9642 |
0.08 |
| chr3_148813708_148813859 | 0.35 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
9196 |
0.3 |
| chr13_22189476_22189663 | 0.35 |
Vmn1r192 |
vomeronasal 1 receptor 192 |
1429 |
0.18 |
| chr8_48535922_48536090 | 0.34 |
Tenm3 |
teneurin transmembrane protein 3 |
19307 |
0.27 |
| chr6_28891682_28891856 | 0.33 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
11726 |
0.21 |
| chr6_140059358_140059541 | 0.33 |
Gm44156 |
predicted gene, 44156 |
1860 |
0.21 |
| chr1_187979615_187979766 | 0.33 |
Esrrg |
estrogen-related receptor gamma |
18137 |
0.23 |
| chr16_43345942_43346096 | 0.33 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
17029 |
0.16 |
| chr17_80677275_80677429 | 0.33 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
26202 |
0.2 |
| chr6_90434918_90435091 | 0.33 |
Cfap100 |
cilia and flagella associated protein 100 |
6207 |
0.12 |
| chr7_50105985_50106173 | 0.33 |
Nell1 |
NEL-like 1 |
7769 |
0.32 |
| chr13_4467812_4467987 | 0.32 |
Gm48010 |
predicted gene, 48010 |
1333 |
0.42 |
| chr7_38016660_38016811 | 0.32 |
Uri1 |
URI1, prefoldin-like chaperone |
2787 |
0.27 |
| chr7_50993814_50993984 | 0.32 |
Gm21036 |
predicted gene, 21036 |
6991 |
0.16 |
| chr9_7891300_7891457 | 0.32 |
Birc3 |
baculoviral IAP repeat-containing 3 |
18192 |
0.17 |
| chr2_167330564_167330838 | 0.32 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
16801 |
0.17 |
| chr15_77072900_77073074 | 0.31 |
Gm24753 |
predicted gene, 24753 |
5196 |
0.12 |
| chr9_113968894_113969045 | 0.31 |
Ubp1 |
upstream binding protein 1 |
256 |
0.9 |
| chr14_79133487_79133687 | 0.31 |
Vwa8 |
von Willebrand factor A domain containing 8 |
10334 |
0.15 |
| chr2_14216800_14216951 | 0.31 |
Mrc1 |
mannose receptor, C type 1 |
12517 |
0.15 |
| chr17_47990768_47990919 | 0.31 |
Gm14871 |
predicted gene 14871 |
12729 |
0.14 |
| chr19_43811537_43811693 | 0.31 |
Abcc2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
1093 |
0.45 |
| chr7_118697788_118697947 | 0.30 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
7513 |
0.13 |
| chr3_84061056_84061215 | 0.30 |
Gm36981 |
predicted gene, 36981 |
2317 |
0.33 |
| chr14_12070865_12071149 | 0.30 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
20153 |
0.22 |
| chr1_23484714_23484885 | 0.30 |
Gm7784 |
predicted gene 7784 |
5260 |
0.27 |
| chr19_33480626_33480777 | 0.29 |
Lipo5 |
lipase, member O5 |
7510 |
0.16 |
| chrX_167880328_167880485 | 0.29 |
Frmpd4 |
FERM and PDZ domain containing 4 |
17152 |
0.21 |
| chr15_3482247_3482450 | 0.29 |
Ghr |
growth hormone receptor |
10704 |
0.28 |
| chr10_87936414_87936806 | 0.29 |
Tyms-ps |
thymidylate synthase, pseudogene |
30237 |
0.15 |
| chr8_33950546_33950701 | 0.29 |
Rbpms |
RNA binding protein gene with multiple splicing |
20760 |
0.14 |
| chr16_77398451_77398621 | 0.29 |
Gm21816 |
predicted gene, 21816 |
1494 |
0.29 |
| chr19_29148038_29148260 | 0.29 |
Gm5518 |
predicted gene 5518 |
11969 |
0.14 |
| chr17_15341574_15341736 | 0.29 |
Gm7423 |
predicted gene 7423 |
19773 |
0.14 |
| chr6_51701361_51701531 | 0.28 |
Gm38811 |
predicted gene, 38811 |
9635 |
0.25 |
| chr11_78567734_78567893 | 0.28 |
Nlk |
nemo like kinase |
1103 |
0.27 |
| chr19_36606384_36606566 | 0.28 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
1308 |
0.49 |
| chr9_83566163_83566322 | 0.28 |
Gm39384 |
predicted gene, 39384 |
7047 |
0.18 |
| chr1_9971330_9971494 | 0.28 |
Tcf24 |
transcription factor 24 |
3480 |
0.12 |
| chr4_102527232_102527683 | 0.28 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
42638 |
0.2 |
| chr19_33489637_33489788 | 0.28 |
Lipo5 |
lipase, member O5 |
16521 |
0.14 |
| chr13_111295092_111295280 | 0.27 |
Actbl2 |
actin, beta-like 2 |
40173 |
0.13 |
| chr3_32530121_32530289 | 0.27 |
Mfn1 |
mitofusin 1 |
666 |
0.62 |
| chr3_138363372_138363558 | 0.27 |
Adh6-ps1 |
alcohol dehydrogenase 6 (class V), pseudogene 1 |
10656 |
0.12 |
| chr7_49393023_49393262 | 0.27 |
Nav2 |
neuron navigator 2 |
28421 |
0.19 |
| chr2_42039219_42039481 | 0.27 |
Gm13461 |
predicted gene 13461 |
50521 |
0.18 |
| chr13_25083206_25083394 | 0.27 |
Dcdc2a |
doublecortin domain containing 2a |
13047 |
0.18 |
| chr2_147873093_147873253 | 0.27 |
Gm25516 |
predicted gene, 25516 |
1705 |
0.48 |
| chr11_99739497_99739661 | 0.27 |
Gm14180 |
predicted gene 14180 |
5289 |
0.06 |
| chr5_90322868_90323041 | 0.27 |
Ankrd17 |
ankyrin repeat domain 17 |
16789 |
0.19 |
| chr10_86350127_86350278 | 0.27 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
47348 |
0.13 |
| chr18_35707850_35708033 | 0.27 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
441 |
0.67 |
| chr10_111576009_111576338 | 0.27 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
18100 |
0.14 |
| chr1_53291339_53291521 | 0.26 |
Pms1 |
PMS1 homolog 1, mismatch repair system component |
5537 |
0.12 |
| chr1_9985468_9985649 | 0.26 |
Ppp1r42 |
protein phosphatase 1, regulatory subunit 42 |
134 |
0.92 |
| chr9_74801197_74801365 | 0.26 |
Gm22315 |
predicted gene, 22315 |
19211 |
0.16 |
| chr5_16316197_16316406 | 0.26 |
Cacna2d1 |
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
9684 |
0.24 |
| chr15_62644818_62645021 | 0.26 |
Gm24810 |
predicted gene, 24810 |
8085 |
0.29 |
| chr18_11858447_11858616 | 0.25 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
14231 |
0.16 |
| chr1_9988282_9988979 | 0.25 |
Ppp1r42 |
protein phosphatase 1, regulatory subunit 42 |
2938 |
0.14 |
| chr2_75770625_75770799 | 0.25 |
Gm13657 |
predicted gene 13657 |
6476 |
0.15 |
| chr2_84357197_84357348 | 0.25 |
Calcrl |
calcitonin receptor-like |
18086 |
0.19 |
| chr16_58663097_58663248 | 0.25 |
Cpox |
coproporphyrinogen oxidase |
7120 |
0.13 |
| chr6_42375567_42375731 | 0.25 |
2010310C07Rik |
RIKEN cDNA 2010310C07 gene |
2307 |
0.14 |
| chr9_122848915_122849207 | 0.25 |
Gm47140 |
predicted gene, 47140 |
643 |
0.55 |
| chr4_15481608_15481775 | 0.25 |
Gm11856 |
predicted gene 11856 |
4675 |
0.28 |
| chr3_107238675_107238837 | 0.25 |
Prok1 |
prokineticin 1 |
104 |
0.95 |
| chr2_68890514_68890665 | 0.25 |
Gm37159 |
predicted gene, 37159 |
777 |
0.58 |
| chr6_72897626_72897819 | 0.24 |
Kcmf1 |
potassium channel modulatory factor 1 |
1361 |
0.34 |
| chr19_8252613_8252782 | 0.24 |
Gm41804 |
predicted gene, 41804 |
729 |
0.61 |
| chr1_164426310_164426497 | 0.24 |
Gm37411 |
predicted gene, 37411 |
1690 |
0.31 |
| chr3_64468587_64468748 | 0.24 |
Vmn2r-ps8 |
vomeronasal 2, receptor, pseudogene 8 |
8351 |
0.13 |
| chr3_108289571_108289732 | 0.24 |
Gm43099 |
predicted gene 43099 |
1770 |
0.18 |
| chr11_50175754_50175938 | 0.24 |
Mrnip |
MRN complex interacting protein |
939 |
0.42 |
| chr5_91200748_91201014 | 0.24 |
Gm23092 |
predicted gene, 23092 |
17123 |
0.2 |
| chr4_102811502_102811684 | 0.24 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
51068 |
0.15 |
| chr2_134826467_134826626 | 0.24 |
Gm14036 |
predicted gene 14036 |
22597 |
0.2 |
| chr3_31376198_31376349 | 0.24 |
Gm38025 |
predicted gene, 38025 |
35261 |
0.18 |
| chr15_53164134_53164470 | 0.24 |
Ext1 |
exostosin glycosyltransferase 1 |
31953 |
0.25 |
| chr12_69419094_69419426 | 0.24 |
Gm9887 |
predicted gene 9887 |
46792 |
0.08 |
| chr4_101393290_101393478 | 0.24 |
Gm12798 |
predicted gene 12798 |
9697 |
0.12 |
| chr13_63665698_63665976 | 0.23 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr10_111171839_111171990 | 0.23 |
Osbpl8 |
oxysterol binding protein-like 8 |
7112 |
0.15 |
| chr10_40246394_40246686 | 0.23 |
Rpf2 |
ribosome production factor 2 homolog |
453 |
0.69 |
| chr6_54032027_54032218 | 0.23 |
Chn2 |
chimerin 2 |
7432 |
0.21 |
| chr15_81521072_81521223 | 0.23 |
Gm5218 |
predicted gene 5218 |
21602 |
0.11 |
| chr7_14307656_14308010 | 0.23 |
Phf20-ps |
PHD finger protein 20, pseudogene |
3053 |
0.2 |
| chr3_149179928_149180099 | 0.23 |
Gm42647 |
predicted gene 42647 |
30258 |
0.19 |
| chr16_92076898_92077091 | 0.23 |
Slc5a3 |
solute carrier family 5 (inositol transporters), member 3 |
1068 |
0.38 |
| chr19_34712180_34712357 | 0.23 |
Gm18425 |
predicted gene, 18425 |
2260 |
0.22 |
| chr16_70331211_70331434 | 0.23 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
17188 |
0.24 |
| chr6_53521285_53521560 | 0.22 |
Creb5 |
cAMP responsive element binding protein 5 |
51878 |
0.15 |
| chr2_42022610_42022815 | 0.22 |
Gm13461 |
predicted gene 13461 |
33883 |
0.23 |
| chr6_8429968_8430158 | 0.22 |
Umad1 |
UMAP1-MVP12 associated (UMA) domain containing 1 |
10385 |
0.16 |
| chr7_80789186_80789337 | 0.22 |
Iqgap1 |
IQ motif containing GTPase activating protein 1 |
14002 |
0.16 |
| chr10_121349498_121349682 | 0.22 |
Gm48435 |
predicted gene, 48435 |
870 |
0.44 |
| chr11_16912365_16912516 | 0.22 |
Egfr |
epidermal growth factor receptor |
7255 |
0.19 |
| chr1_73684327_73684549 | 0.22 |
6030407O03Rik |
RIKEN cDNA 6030407O03 gene |
59509 |
0.12 |
| chr3_18157205_18157356 | 0.22 |
Gm23686 |
predicted gene, 23686 |
20345 |
0.21 |
| chr15_38914094_38914399 | 0.22 |
Baalc |
brain and acute leukemia, cytoplasmic |
18898 |
0.16 |
| chr11_16863303_16863761 | 0.22 |
Egfr |
epidermal growth factor receptor |
14618 |
0.19 |
| chr1_67228479_67228681 | 0.22 |
Gm15668 |
predicted gene 15668 |
20620 |
0.2 |
| chr17_86647655_86647806 | 0.21 |
Gm18832 |
predicted gene, 18832 |
37766 |
0.16 |
| chr1_75337875_75338132 | 0.21 |
Dnpep |
aspartyl aminopeptidase |
20013 |
0.09 |
| chr3_18148711_18148862 | 0.21 |
Gm23686 |
predicted gene, 23686 |
28839 |
0.19 |
| chr7_112688480_112688662 | 0.21 |
Tead1 |
TEA domain family member 1 |
3127 |
0.21 |
| chr17_65788655_65788828 | 0.21 |
Ppp4r1 |
protein phosphatase 4, regulatory subunit 1 |
5142 |
0.19 |
| chr6_111600197_111600355 | 0.21 |
Gm22093 |
predicted gene, 22093 |
2976 |
0.4 |
| chr3_18148886_18149070 | 0.21 |
Gm23686 |
predicted gene, 23686 |
28647 |
0.19 |
| chr3_27905130_27905320 | 0.21 |
Tmem212 |
transmembrane protein 212 |
8857 |
0.22 |
| chr4_130704823_130704974 | 0.21 |
Pum1 |
pumilio RNA-binding family member 1 |
41492 |
0.1 |
| chr1_184053978_184054158 | 0.21 |
Dusp10 |
dual specificity phosphatase 10 |
19687 |
0.21 |
| chr13_80887789_80888455 | 0.21 |
Arrdc3 |
arrestin domain containing 3 |
1982 |
0.27 |
| chr1_71723871_71724224 | 0.21 |
Gm5256 |
predicted gene 5256 |
9123 |
0.19 |
| chr14_26432359_26432703 | 0.21 |
Slmap |
sarcolemma associated protein |
2430 |
0.24 |
| chr5_74833941_74834092 | 0.21 |
Gm17906 |
predicted gene, 17906 |
49319 |
0.12 |
| chr10_81388466_81388771 | 0.21 |
Gm48552 |
predicted gene, 48552 |
387 |
0.55 |
| chr3_130409662_130410071 | 0.21 |
Col25a1 |
collagen, type XXV, alpha 1 |
68950 |
0.11 |
| chr1_184100523_184100685 | 0.21 |
Dusp10 |
dual specificity phosphatase 10 |
66223 |
0.12 |
| chr4_99068687_99068838 | 0.21 |
Dock7 |
dedicator of cytokinesis 7 |
1651 |
0.35 |
| chr16_56038236_56038391 | 0.21 |
Trmt10c |
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
494 |
0.65 |
| chr11_5306963_5307114 | 0.20 |
Gm30172 |
predicted gene, 30172 |
32953 |
0.14 |
| chr14_30184944_30185295 | 0.20 |
Cacna1d |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
9728 |
0.2 |
| chr1_185216852_185217011 | 0.20 |
Rab3gap2 |
RAB3 GTPase activating protein subunit 2 |
12753 |
0.13 |
| chr13_34465973_34466145 | 0.20 |
Gm47120 |
predicted gene, 47120 |
39628 |
0.12 |
| chr1_43935708_43935875 | 0.20 |
Tpp2 |
tripeptidyl peptidase II |
1746 |
0.27 |
| chr7_28927529_28927703 | 0.20 |
Actn4 |
actinin alpha 4 |
8395 |
0.1 |
| chr5_90520250_90520471 | 0.20 |
Afm |
afamin |
1411 |
0.31 |
| chr9_7858246_7858397 | 0.20 |
Birc3 |
baculoviral IAP repeat-containing 3 |
2005 |
0.3 |
| chr5_28060446_28060778 | 0.20 |
Gm26608 |
predicted gene, 26608 |
5153 |
0.18 |
| chr7_145092850_145093049 | 0.20 |
Gm45181 |
predicted gene 45181 |
70047 |
0.1 |
| chr1_186952519_186952695 | 0.20 |
Gm3809 |
predicted gene 3809 |
7901 |
0.15 |
| chr8_80046818_80046969 | 0.20 |
Hhip |
Hedgehog-interacting protein |
11113 |
0.21 |
| chrX_98051624_98051802 | 0.20 |
Gm14802 |
predicted gene 14802 |
11338 |
0.3 |
| chr7_14438643_14438794 | 0.20 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
157 |
0.95 |
| chr12_12510893_12511142 | 0.20 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
118402 |
0.06 |
| chr11_90643436_90643587 | 0.20 |
Cox11 |
cytochrome c oxidase assembly protein 11, copper chaperone |
3333 |
0.22 |
| chr12_71399037_71399230 | 0.20 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
14925 |
0.18 |
| chr1_88048809_88049389 | 0.20 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6289 |
0.08 |
| chr3_138242749_138243005 | 0.19 |
0610031O16Rik |
RIKEN cDNA 0610031O16 gene |
2472 |
0.18 |
| chr17_47627069_47627220 | 0.19 |
Usp49 |
ubiquitin specific peptidase 49 |
3546 |
0.11 |
| chr18_37825326_37825501 | 0.19 |
Gm38182 |
predicted gene, 38182 |
676 |
0.4 |
| chr8_91529579_91530083 | 0.19 |
Gm45290 |
predicted gene 45290 |
2893 |
0.21 |
| chr6_35253418_35253775 | 0.19 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
864 |
0.54 |
| chr1_184330776_184331172 | 0.19 |
Gm37223 |
predicted gene, 37223 |
27355 |
0.2 |
| chr14_36743243_36743409 | 0.19 |
Gm25204 |
predicted gene, 25204 |
24099 |
0.23 |
| chr5_23562685_23562882 | 0.19 |
Srpk2 |
serine/arginine-rich protein specific kinase 2 |
13413 |
0.19 |
| chr4_150434207_150434380 | 0.19 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
27899 |
0.2 |
| chr11_87713864_87714038 | 0.19 |
1110028F11Rik |
RIKEN cDNA 1110028F11 gene |
11828 |
0.09 |
| chr2_3415817_3416004 | 0.19 |
Meig1 |
meiosis expressed gene 1 |
2671 |
0.18 |
| chr12_57444501_57444678 | 0.19 |
Gm16246 |
predicted gene 16246 |
5531 |
0.24 |
| chr13_36053796_36053967 | 0.19 |
Gm48770 |
predicted gene, 48770 |
58428 |
0.08 |
| chr1_7096771_7096931 | 0.19 |
Pcmtd1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
7685 |
0.17 |
| chr3_4950385_4950550 | 0.19 |
Gm6140 |
predicted gene 6140 |
59236 |
0.14 |
| chr9_107674673_107674872 | 0.19 |
Gnat1 |
guanine nucleotide binding protein, alpha transducing 1 |
1669 |
0.18 |
| chr13_9145204_9145368 | 0.19 |
Gm28155 |
predicted gene 28155 |
4405 |
0.19 |
| chr4_82404190_82404364 | 0.19 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
35133 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.0 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |