Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
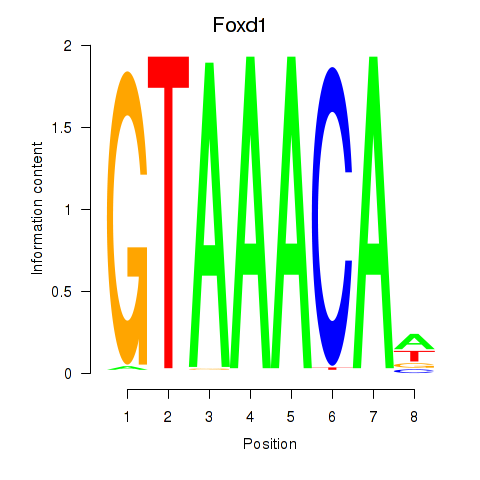
Results for Foxd1
Z-value: 1.36
Transcription factors associated with Foxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd1
|
ENSMUSG00000078302.3 | forkhead box D1 |
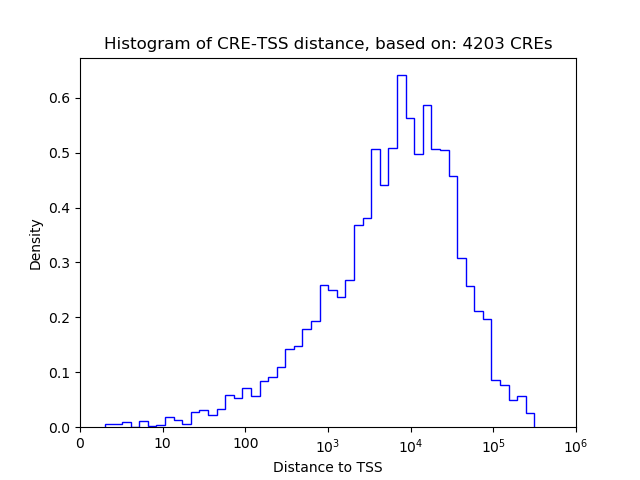
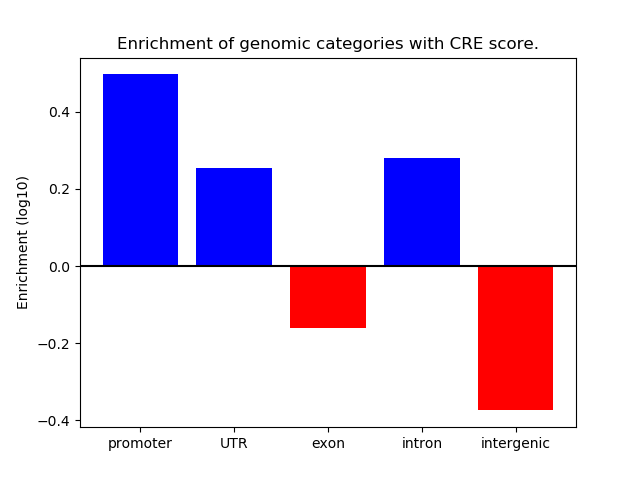
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_98352948_98353139 | Foxd1 | 1199 | 0.402992 | 0.88 | 2.2e-02 | Click! |
Activity of the Foxd1 motif across conditions
Conditions sorted by the z-value of the Foxd1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
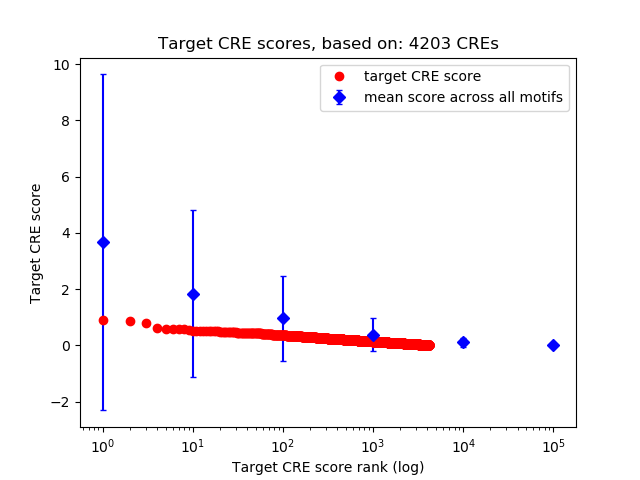
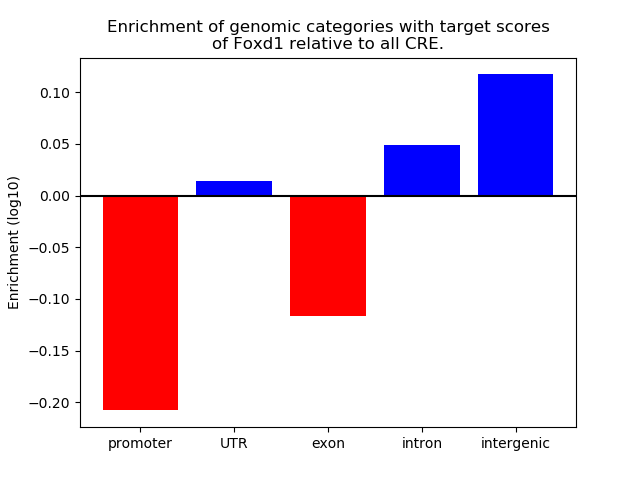
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_117342819_117343008 | 0.89 |
Rnf220 |
ring finger protein 220 |
32544 |
0.12 |
| chr10_8092375_8092549 | 0.87 |
Gm48614 |
predicted gene, 48614 |
71170 |
0.1 |
| chr5_134941937_134942100 | 0.81 |
Mettl27 |
methyltransferase like 27 |
1556 |
0.18 |
| chrX_105934496_105934654 | 0.62 |
Atrx |
ATRX, chromatin remodeler |
5172 |
0.21 |
| chr12_82743501_82743658 | 0.59 |
Gm22149 |
predicted gene, 22149 |
28877 |
0.22 |
| chr3_101881841_101882004 | 0.58 |
Slc22a15 |
solute carrier family 22 (organic anion/cation transporter), member 15 |
4035 |
0.26 |
| chr19_32685579_32685730 | 0.57 |
Atad1 |
ATPase family, AAA domain containing 1 |
9216 |
0.24 |
| chr19_32282556_32283119 | 0.56 |
Sgms1 |
sphingomyelin synthase 1 |
5873 |
0.26 |
| chr13_77864889_77865040 | 0.54 |
Fam172a |
family with sequence similarity 172, member A |
156180 |
0.04 |
| chr1_179833675_179833847 | 0.53 |
Ahctf1 |
AT hook containing transcription factor 1 |
30081 |
0.16 |
| chr10_8086797_8087175 | 0.53 |
Gm48614 |
predicted gene, 48614 |
65694 |
0.11 |
| chr12_62770911_62771078 | 0.53 |
Tspyl-ps |
testis-specific protein, Y-encoded-like, pseudogene |
66718 |
0.1 |
| chr5_134550893_134551073 | 0.52 |
Gm42884 |
predicted gene 42884 |
1074 |
0.29 |
| chr7_122665413_122665987 | 0.52 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
4792 |
0.21 |
| chr16_17217236_17217387 | 0.52 |
Rimbp3 |
RIMS binding protein 3 |
8708 |
0.09 |
| chr11_57983626_57984029 | 0.52 |
Gm12249 |
predicted gene 12249 |
10693 |
0.15 |
| chr16_22694632_22694783 | 0.52 |
Gm8118 |
predicted gene 8118 |
8513 |
0.18 |
| chr13_60267432_60267612 | 0.51 |
Gm24999 |
predicted gene, 24999 |
26150 |
0.16 |
| chr12_88472970_88473131 | 0.50 |
Gm48642 |
predicted gene, 48642 |
30718 |
0.1 |
| chr15_25914259_25914426 | 0.49 |
Retreg1 |
reticulophagy regulator 1 |
3918 |
0.25 |
| chr10_59537699_59537863 | 0.47 |
Gm10322 |
predicted gene 10322 |
78282 |
0.08 |
| chr3_83996881_83997402 | 0.47 |
Tmem131l |
transmembrane 131 like |
28913 |
0.2 |
| chr12_82353794_82353959 | 0.47 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
11609 |
0.28 |
| chr15_55819504_55819678 | 0.47 |
Sntb1 |
syntrophin, basic 1 |
86719 |
0.08 |
| chr14_28652150_28652318 | 0.47 |
Gm35164 |
predicted gene, 35164 |
2475 |
0.38 |
| chr10_37711570_37711751 | 0.46 |
Gm24710 |
predicted gene, 24710 |
220020 |
0.02 |
| chr5_63893264_63893450 | 0.46 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
15881 |
0.17 |
| chr10_122361947_122362104 | 0.46 |
Gm36041 |
predicted gene, 36041 |
24867 |
0.2 |
| chr15_7088669_7089236 | 0.46 |
Lifr |
LIF receptor alpha |
1662 |
0.5 |
| chr10_94568025_94568184 | 0.46 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
7153 |
0.17 |
| chr19_36627452_36627789 | 0.46 |
Hectd2os |
Hectd2, opposite strand |
1596 |
0.43 |
| chr10_28524334_28524485 | 0.45 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
35742 |
0.22 |
| chrX_106036704_106036855 | 0.45 |
Atp7a |
ATPase, Cu++ transporting, alpha polypeptide |
9457 |
0.17 |
| chr4_41630291_41630458 | 0.44 |
Dnaic1 |
dynein, axonemal, intermediate chain 1 |
1270 |
0.29 |
| chr18_60748821_60749236 | 0.44 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
378 |
0.77 |
| chr8_94678714_94678865 | 0.44 |
Pllp |
plasma membrane proteolipid |
183 |
0.91 |
| chr8_11148911_11149083 | 0.44 |
Gm44717 |
predicted gene 44717 |
249 |
0.91 |
| chr2_103870054_103870389 | 0.44 |
Gm13876 |
predicted gene 13876 |
18103 |
0.09 |
| chr14_105603866_105604023 | 0.44 |
9330188P03Rik |
RIKEN cDNA 9330188P03 gene |
13887 |
0.16 |
| chr15_67420967_67421299 | 0.44 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
194364 |
0.03 |
| chrX_164035471_164035631 | 0.44 |
Car5b |
carbonic anhydrase 5b, mitochondrial |
7554 |
0.2 |
| chr18_44610487_44610664 | 0.44 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
51090 |
0.14 |
| chr2_24742080_24742251 | 0.44 |
Cacna1b |
calcium channel, voltage-dependent, N type, alpha 1B subunit |
20882 |
0.16 |
| chr9_41306336_41306516 | 0.43 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
20651 |
0.16 |
| chr5_65096928_65097079 | 0.43 |
Tmem156 |
transmembrane protein 156 |
4871 |
0.18 |
| chr1_86479051_86479408 | 0.43 |
Rpl30-ps6 |
ribosomal protein L30, pseudogene 6 |
5570 |
0.15 |
| chr13_19306748_19306899 | 0.43 |
Trgc3 |
T cell receptor gamma, constant 3 |
45939 |
0.13 |
| chr6_48857733_48857900 | 0.43 |
Gm7932 |
predicted gene 7932 |
2513 |
0.13 |
| chr12_34406620_34406775 | 0.43 |
Hdac9 |
histone deacetylase 9 |
30610 |
0.24 |
| chr2_84180208_84180377 | 0.43 |
Gm25972 |
predicted gene, 25972 |
30237 |
0.16 |
| chr18_11858447_11858616 | 0.43 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
14231 |
0.16 |
| chr1_167320339_167320493 | 0.43 |
Tmco1 |
transmembrane and coiled-coil domains 1 |
6279 |
0.12 |
| chr18_46712101_46712569 | 0.42 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
15694 |
0.12 |
| chr1_133265093_133265439 | 0.42 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
4190 |
0.17 |
| chr15_96372725_96372895 | 0.42 |
Arid2 |
AT rich interactive domain 2 (ARID, RFX-like) |
6004 |
0.21 |
| chr6_87428535_87428790 | 0.42 |
Bmp10 |
bone morphogenetic protein 10 |
332 |
0.85 |
| chr9_105878487_105878719 | 0.42 |
Col6a5 |
collagen, type VI, alpha 5 |
3203 |
0.27 |
| chr6_111600197_111600355 | 0.42 |
Gm22093 |
predicted gene, 22093 |
2976 |
0.4 |
| chr4_150825989_150826153 | 0.41 |
Gm13049 |
predicted gene 13049 |
338 |
0.86 |
| chr16_78254868_78255025 | 0.41 |
E330011O21Rik |
RIKEN cDNA E330011O21 gene |
410 |
0.81 |
| chr19_36629491_36629794 | 0.41 |
Hectd2os |
Hectd2, opposite strand |
3618 |
0.27 |
| chr6_5496582_5496768 | 0.41 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
366 |
0.92 |
| chr8_4193425_4193618 | 0.40 |
Evi5l |
ecotropic viral integration site 5 like |
2229 |
0.17 |
| chr11_3161350_3161520 | 0.40 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
3162 |
0.16 |
| chr5_52360112_52360263 | 0.40 |
Sod3 |
superoxide dismutase 3, extracellular |
3604 |
0.19 |
| chr4_44994891_44995176 | 0.40 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
13533 |
0.1 |
| chr10_63413324_63413532 | 0.40 |
Gm7530 |
predicted gene 7530 |
429 |
0.77 |
| chr6_126033747_126033913 | 0.40 |
Ntf3 |
neurotrophin 3 |
131130 |
0.05 |
| chr10_95596327_95596490 | 0.40 |
Nudt4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
32262 |
0.1 |
| chr1_24693656_24693810 | 0.40 |
Lmbrd1 |
LMBR1 domain containing 1 |
14807 |
0.17 |
| chr1_165609107_165609258 | 0.39 |
Mpzl1 |
myelin protein zero-like 1 |
364 |
0.78 |
| chr11_82803141_82803324 | 0.39 |
Lig3 |
ligase III, DNA, ATP-dependent |
4883 |
0.12 |
| chr15_77891897_77892048 | 0.39 |
Txn2 |
thioredoxin 2 |
23728 |
0.13 |
| chr5_104086712_104086871 | 0.39 |
Gm17660 |
predicted gene, 17660 |
9183 |
0.11 |
| chr7_84201215_84201396 | 0.39 |
Gm44826 |
predicted gene 44826 |
9209 |
0.15 |
| chr7_80402754_80402960 | 0.38 |
Furin |
furin (paired basic amino acid cleaving enzyme) |
95 |
0.93 |
| chr4_155245715_155246079 | 0.38 |
Faap20 |
Fanconi anemia core complex associated protein 20 |
3905 |
0.19 |
| chr16_33892287_33892604 | 0.38 |
Itgb5 |
integrin beta 5 |
498 |
0.81 |
| chr14_27310963_27311149 | 0.38 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
25010 |
0.19 |
| chr3_65859331_65859533 | 0.38 |
Gm37131 |
predicted gene, 37131 |
5397 |
0.18 |
| chr16_22439730_22439912 | 0.38 |
Etv5 |
ets variant 5 |
102 |
0.97 |
| chr6_17513498_17513674 | 0.38 |
Met |
met proto-oncogene |
21415 |
0.21 |
| chr2_152817536_152817750 | 0.38 |
Bcl2l1 |
BCL2-like 1 |
10892 |
0.12 |
| chr10_116287055_116287225 | 0.38 |
Gm47625 |
predicted gene, 47625 |
2591 |
0.27 |
| chr1_86447982_86448140 | 0.37 |
Tex44 |
testis expressed 44 |
21732 |
0.1 |
| chr13_41321516_41321722 | 0.37 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
37628 |
0.1 |
| chr10_61327652_61327821 | 0.37 |
Pald1 |
phosphatase domain containing, paladin 1 |
14591 |
0.12 |
| chr4_126113392_126113559 | 0.37 |
Stk40 |
serine/threonine kinase 40 |
4797 |
0.12 |
| chr16_91146000_91146151 | 0.37 |
Gm49612 |
predicted gene, 49612 |
10599 |
0.1 |
| chr17_86142116_86142294 | 0.37 |
Srbd1 |
S1 RNA binding domain 1 |
2970 |
0.28 |
| chr5_46856306_46856720 | 0.37 |
Gm43092 |
predicted gene 43092 |
61833 |
0.16 |
| chr2_72880103_72880418 | 0.37 |
Sp3 |
trans-acting transcription factor 3 |
60824 |
0.09 |
| chr10_69930149_69930514 | 0.36 |
Ank3 |
ankyrin 3, epithelial |
4198 |
0.35 |
| chr12_113110325_113110520 | 0.36 |
Mta1 |
metastasis associated 1 |
12021 |
0.1 |
| chr12_108304077_108304251 | 0.36 |
Hhipl1 |
hedgehog interacting protein-like 1 |
2106 |
0.27 |
| chr19_4800564_4800742 | 0.36 |
Gm21844 |
predicted gene, 21844 |
859 |
0.32 |
| chr18_38521952_38522103 | 0.36 |
Gm50349 |
predicted gene, 50349 |
40724 |
0.12 |
| chr14_54434127_54434453 | 0.36 |
Mmp14 |
matrix metallopeptidase 14 (membrane-inserted) |
1854 |
0.16 |
| chr7_134497342_134497526 | 0.36 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
1550 |
0.55 |
| chr12_21166146_21166319 | 0.35 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
54278 |
0.1 |
| chr15_97590730_97591033 | 0.35 |
Gm49506 |
predicted gene, 49506 |
32591 |
0.19 |
| chr1_75521864_75522062 | 0.35 |
Stk11ip |
serine/threonine kinase 11 interacting protein |
152 |
0.9 |
| chr9_119342839_119343002 | 0.35 |
Acaa1a |
acetyl-Coenzyme A acyltransferase 1A |
147 |
0.91 |
| chr10_62795188_62795339 | 0.35 |
Ccar1 |
cell division cycle and apoptosis regulator 1 |
2977 |
0.15 |
| chr10_127653471_127653627 | 0.35 |
Stat6 |
signal transducer and activator of transcription 6 |
470 |
0.63 |
| chr16_35949490_35949675 | 0.35 |
Parp9 |
poly (ADP-ribose) polymerase family, member 9 |
1298 |
0.3 |
| chr8_84740975_84741146 | 0.35 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
18053 |
0.1 |
| chr6_97971154_97971321 | 0.35 |
Mitf |
melanogenesis associated transcription factor |
20555 |
0.25 |
| chr15_66911047_66911248 | 0.35 |
Ccn4 |
cellular communication network factor 4 |
1836 |
0.32 |
| chr9_48592438_48592815 | 0.35 |
Nnmt |
nicotinamide N-methyltransferase |
4749 |
0.26 |
| chr15_59391938_59392317 | 0.35 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
17840 |
0.18 |
| chr10_11150027_11150209 | 0.35 |
Shprh |
SNF2 histone linker PHD RING helicase |
594 |
0.78 |
| chr1_165849593_165849764 | 0.35 |
Gm37469 |
predicted gene, 37469 |
634 |
0.54 |
| chr4_117852028_117852199 | 0.35 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
2743 |
0.15 |
| chr7_138867340_138867520 | 0.34 |
Ppp2r2d |
protein phosphatase 2, regulatory subunit B, delta |
980 |
0.43 |
| chr14_46017259_46017572 | 0.34 |
Gm6580 |
predicted gene 6580 |
10013 |
0.17 |
| chr10_8228115_8228580 | 0.34 |
Gm30906 |
predicted gene, 30906 |
52216 |
0.15 |
| chr11_44155690_44155841 | 0.34 |
Gm12154 |
predicted gene 12154 |
111444 |
0.06 |
| chrX_112984511_112984736 | 0.34 |
Gm14938 |
predicted gene 14938 |
17534 |
0.15 |
| chrX_129949506_129949684 | 0.34 |
Diaph2 |
diaphanous related formin 2 |
22938 |
0.28 |
| chr8_10904045_10904209 | 0.34 |
Gm2814 |
predicted gene 2814 |
3569 |
0.12 |
| chr6_145767466_145768203 | 0.34 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
17263 |
0.18 |
| chr5_3488489_3488661 | 0.34 |
Gm17590 |
predicted gene, 17590 |
7702 |
0.13 |
| chr5_113204337_113204488 | 0.33 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
16824 |
0.12 |
| chr7_67951889_67952040 | 0.33 |
Igf1r |
insulin-like growth factor I receptor |
863 |
0.66 |
| chr5_114796772_114796944 | 0.33 |
Ankrd13a |
ankyrin repeat domain 13a |
1115 |
0.31 |
| chr6_35253418_35253775 | 0.33 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
864 |
0.54 |
| chr8_121580222_121580383 | 0.33 |
Gm17786 |
predicted gene, 17786 |
1463 |
0.23 |
| chr2_27215557_27215850 | 0.33 |
Sardh |
sarcosine dehydrogenase |
71 |
0.96 |
| chr16_18430516_18430667 | 0.33 |
Txnrd2 |
thioredoxin reductase 2 |
1666 |
0.22 |
| chr3_122565390_122565553 | 0.33 |
Gm43569 |
predicted gene 43569 |
421 |
0.72 |
| chr15_67423761_67424241 | 0.33 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
197232 |
0.03 |
| chr10_24087275_24087474 | 0.33 |
Taar8b |
trace amine-associated receptor 8B |
4920 |
0.1 |
| chr5_66798803_66798961 | 0.33 |
Limch1 |
LIM and calponin homology domains 1 |
52993 |
0.11 |
| chr14_35146195_35146544 | 0.33 |
Gm49034 |
predicted gene, 49034 |
73069 |
0.12 |
| chr11_94365374_94365707 | 0.33 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
2383 |
0.24 |
| chr12_76271569_76271743 | 0.33 |
Gm47526 |
predicted gene, 47526 |
6060 |
0.12 |
| chr6_16701972_16702148 | 0.33 |
Gm36669 |
predicted gene, 36669 |
75464 |
0.11 |
| chr3_122417274_122417702 | 0.33 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
2269 |
0.24 |
| chr12_79140897_79141048 | 0.33 |
Arg2 |
arginase type II |
10195 |
0.09 |
| chr9_57281733_57281969 | 0.33 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
19239 |
0.14 |
| chr17_50468359_50468519 | 0.32 |
Plcl2 |
phospholipase C-like 2 |
40964 |
0.19 |
| chr11_50622338_50622503 | 0.32 |
Gm12198 |
predicted gene 12198 |
19972 |
0.17 |
| chr9_61151961_61152206 | 0.32 |
I730028E13Rik |
RIKEN cDNA I730028E13 gene |
13568 |
0.15 |
| chr4_130116362_130116530 | 0.32 |
Pef1 |
penta-EF hand domain containing 1 |
8890 |
0.14 |
| chr18_70611236_70611413 | 0.32 |
Mbd2 |
methyl-CpG binding domain protein 2 |
6449 |
0.21 |
| chr5_146967227_146967378 | 0.32 |
Mtif3 |
mitochondrial translational initiation factor 3 |
3502 |
0.21 |
| chr13_34301223_34301392 | 0.32 |
Gm47086 |
predicted gene, 47086 |
17501 |
0.18 |
| chrX_42112143_42112724 | 0.32 |
Gm14615 |
predicted gene 14615 |
18290 |
0.16 |
| chr9_43561805_43561964 | 0.32 |
Gm36855 |
predicted gene, 36855 |
40880 |
0.13 |
| chr4_86777161_86777312 | 0.32 |
Dennd4c |
DENN/MADD domain containing 4C |
607 |
0.77 |
| chr1_39105614_39105798 | 0.32 |
Gm37091 |
predicted gene, 37091 |
22368 |
0.17 |
| chr16_30603170_30603355 | 0.32 |
Fam43a |
family with sequence similarity 43, member A |
3539 |
0.24 |
| chr10_41636277_41636445 | 0.32 |
Ccdc162 |
coiled-coil domain containing 162 |
23524 |
0.13 |
| chr13_120025614_120025791 | 0.32 |
Gm36079 |
predicted gene, 36079 |
1309 |
0.25 |
| chr15_6709205_6709602 | 0.32 |
Rictor |
RPTOR independent companion of MTOR, complex 2 |
1020 |
0.58 |
| chr5_22255477_22255648 | 0.32 |
Gm16113 |
predicted gene 16113 |
26020 |
0.16 |
| chr12_76009151_76009311 | 0.32 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
13069 |
0.25 |
| chr5_148684126_148684386 | 0.31 |
Gm36186 |
predicted gene, 36186 |
27210 |
0.15 |
| chr2_25363590_25363748 | 0.31 |
Uap1l1 |
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
29 |
0.93 |
| chr3_108083014_108083206 | 0.31 |
Ampd2 |
adenosine monophosphate deaminase 2 |
2285 |
0.13 |
| chr12_71719042_71719430 | 0.31 |
Gm47555 |
predicted gene, 47555 |
35149 |
0.17 |
| chr1_157073744_157073917 | 0.31 |
Gm28694 |
predicted gene 28694 |
19796 |
0.15 |
| chr15_89377307_89377495 | 0.31 |
Tymp |
thymidine phosphorylase |
362 |
0.6 |
| chr11_78418219_78418374 | 0.31 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
3921 |
0.12 |
| chr4_33262151_33262376 | 0.31 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
13753 |
0.16 |
| chr11_7198754_7198905 | 0.31 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1047 |
0.5 |
| chr13_35023472_35023629 | 0.31 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
3544 |
0.15 |
| chr6_5517592_5517751 | 0.31 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
21362 |
0.26 |
| chr7_132931708_132932726 | 0.30 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
1020 |
0.34 |
| chr4_123108028_123108191 | 0.30 |
Bmp8b |
bone morphogenetic protein 8b |
2944 |
0.16 |
| chr10_12278119_12278297 | 0.30 |
Gm48723 |
predicted gene, 48723 |
46453 |
0.13 |
| chr5_145880032_145880197 | 0.30 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
150 |
0.95 |
| chrX_165191272_165191423 | 0.30 |
Glra2 |
glycine receptor, alpha 2 subunit |
136046 |
0.05 |
| chr19_45445850_45446001 | 0.30 |
Btrc |
beta-transducin repeat containing protein |
417 |
0.83 |
| chr8_109993075_109993269 | 0.30 |
Tat |
tyrosine aminotransferase |
2666 |
0.18 |
| chr5_147894276_147894456 | 0.30 |
Slc46a3 |
solute carrier family 46, member 3 |
449 |
0.8 |
| chr5_115167416_115167584 | 0.30 |
Mlec |
malectin |
9321 |
0.1 |
| chr10_118581468_118581731 | 0.30 |
Ifngas1 |
Ifng antisense RNA 1 |
25074 |
0.2 |
| chr10_96646651_96646843 | 0.30 |
Btg1 |
BTG anti-proliferation factor 1 |
29193 |
0.17 |
| chr11_117970308_117970484 | 0.30 |
Socs3 |
suppressor of cytokine signaling 3 |
349 |
0.83 |
| chr2_164445108_164445423 | 0.30 |
Sdc4 |
syndecan 4 |
1378 |
0.23 |
| chr7_70788098_70788301 | 0.30 |
Gm24880 |
predicted gene, 24880 |
35783 |
0.18 |
| chr10_68444703_68445037 | 0.30 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
80897 |
0.09 |
| chr12_113252720_113252871 | 0.30 |
Gm25622 |
predicted gene, 25622 |
38646 |
0.1 |
| chr15_66073884_66074203 | 0.30 |
Gm27153 |
predicted gene 27153 |
509 |
0.83 |
| chr2_156763206_156763372 | 0.30 |
Dlgap4 |
DLG associated protein 4 |
3344 |
0.16 |
| chr9_35134057_35134256 | 0.29 |
Gm24262 |
predicted gene, 24262 |
10480 |
0.12 |
| chr2_50625473_50625690 | 0.29 |
Gm13484 |
predicted gene 13484 |
32486 |
0.2 |
| chrX_100447058_100447240 | 0.29 |
Awat2 |
acyl-CoA wax alcohol acyltransferase 2 |
4432 |
0.18 |
| chr17_12975251_12975429 | 0.29 |
Wtap |
Wilms tumour 1-associating protein |
8133 |
0.09 |
| chr5_63936466_63936784 | 0.29 |
Rell1 |
RELT-like 1 |
5711 |
0.18 |
| chr8_114541036_114541418 | 0.29 |
Gm16116 |
predicted gene 16116 |
53673 |
0.14 |
| chr19_29268750_29269067 | 0.29 |
Jak2 |
Janus kinase 2 |
16467 |
0.17 |
| chr13_115014976_115015141 | 0.29 |
Gm10734 |
predicted gene 10734 |
74443 |
0.07 |
| chr12_110020945_110021120 | 0.29 |
Gm34667 |
predicted gene, 34667 |
2841 |
0.19 |
| chr7_112497046_112497197 | 0.29 |
Parva |
parvin, alpha |
22580 |
0.2 |
| chr14_121315574_121315760 | 0.29 |
Stk24 |
serine/threonine kinase 24 |
12192 |
0.21 |
| chr1_184330776_184331172 | 0.29 |
Gm37223 |
predicted gene, 37223 |
27355 |
0.2 |
| chr5_118291975_118292126 | 0.29 |
Gm25076 |
predicted gene, 25076 |
25601 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.4 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.2 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.0 | GO:0003097 | renal water transport(GO:0003097) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.2 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) |
| 0.0 | 0.0 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.0 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.0 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.0 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.0 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.5 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.3 | GO:0043910 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0018640 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0043823 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0043820 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.3 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |