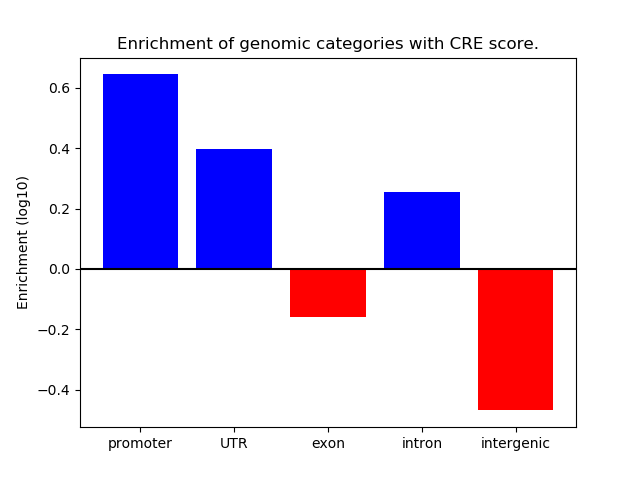
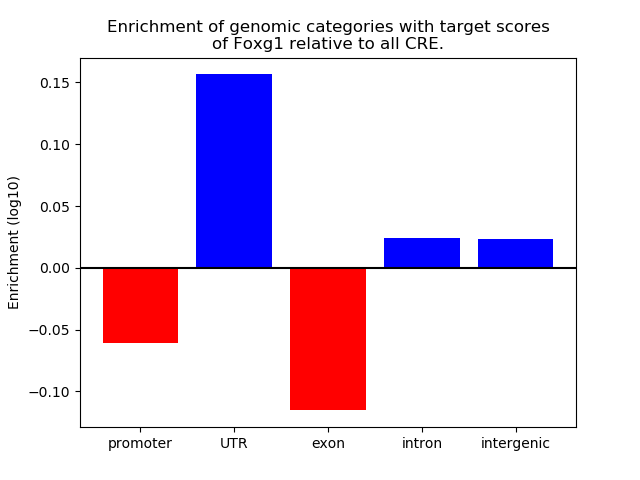
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
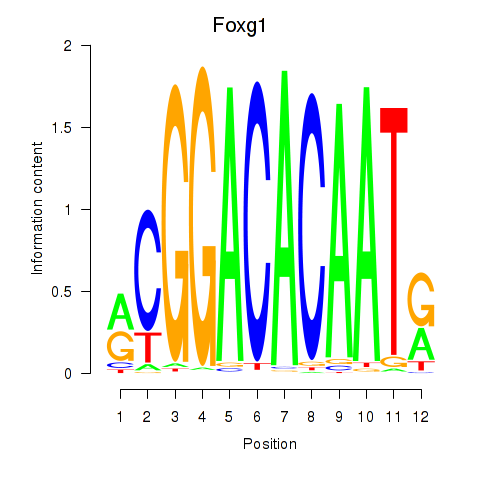
Results for Foxg1
Z-value: 0.91
Transcription factors associated with Foxg1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxg1
|
ENSMUSG00000020950.9 | forkhead box G1 |
Activity of the Foxg1 motif across conditions
Conditions sorted by the z-value of the Foxg1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
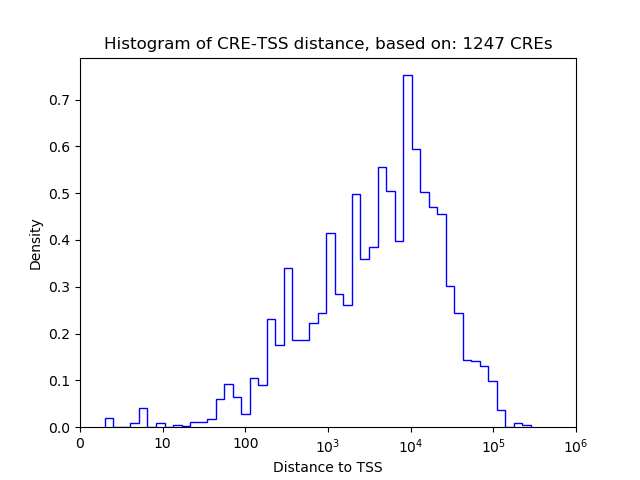
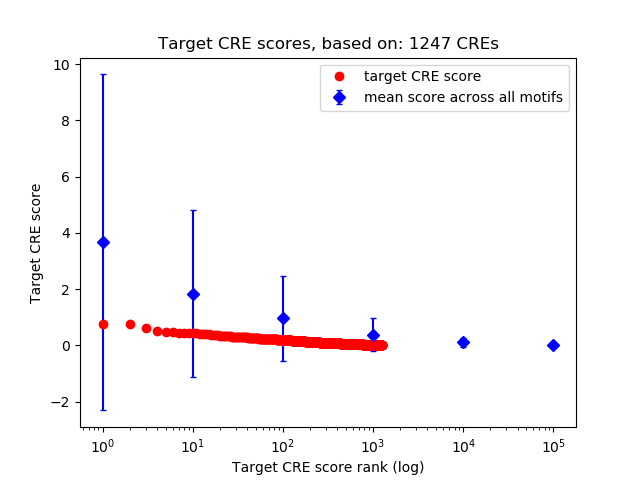
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_141432376_141432556 | 0.75 |
Gm43958 |
predicted gene, 43958 |
9894 |
0.26 |
| chr4_140856184_140856335 | 0.74 |
Padi1 |
peptidyl arginine deiminase, type I |
10481 |
0.12 |
| chr10_61720415_61720605 | 0.61 |
Aifm2 |
apoptosis-inducing factor, mitochondrion-associated 2 |
139 |
0.94 |
| chr9_119148992_119149143 | 0.50 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
996 |
0.4 |
| chr1_189916522_189916681 | 0.49 |
Gm38245 |
predicted gene, 38245 |
642 |
0.69 |
| chr7_119965208_119965675 | 0.48 |
Dnah3 |
dynein, axonemal, heavy chain 3 |
2401 |
0.24 |
| chr10_24835678_24835882 | 0.45 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
333 |
0.88 |
| chr13_58197190_58197341 | 0.44 |
Gm48357 |
predicted gene, 48357 |
5947 |
0.11 |
| chr7_70791296_70791455 | 0.43 |
Gm24880 |
predicted gene, 24880 |
38959 |
0.17 |
| chr5_114154896_114155238 | 0.43 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
8532 |
0.12 |
| chr11_100931218_100931481 | 0.43 |
Stat3 |
signal transducer and activator of transcription 3 |
8031 |
0.14 |
| chr1_166000310_166001131 | 0.42 |
Pou2f1 |
POU domain, class 2, transcription factor 1 |
1889 |
0.25 |
| chr4_33033624_33033916 | 0.41 |
Ube2j1 |
ubiquitin-conjugating enzyme E2J 1 |
2242 |
0.18 |
| chr6_57543051_57543249 | 0.40 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
7682 |
0.16 |
| chr7_80666924_80667352 | 0.39 |
Crtc3 |
CREB regulated transcription coactivator 3 |
21739 |
0.15 |
| chr7_81110178_81110363 | 0.38 |
Slc28a1 |
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
4529 |
0.17 |
| chr18_9078341_9078508 | 0.36 |
Gm18526 |
predicted gene, 18526 |
21831 |
0.24 |
| chr9_30937150_30937309 | 0.36 |
Adamts8 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
5333 |
0.21 |
| chr9_110985950_110986443 | 0.35 |
1700061E17Rik |
RIKEN cDNA 1700061E17 gene |
5 |
0.94 |
| chr5_66067953_66068113 | 0.35 |
Gm43775 |
predicted gene 43775 |
4862 |
0.14 |
| chr11_120815283_120815743 | 0.34 |
Fasn |
fatty acid synthase |
58 |
0.94 |
| chr4_135516112_135516333 | 0.33 |
Gm25317 |
predicted gene, 25317 |
3743 |
0.16 |
| chr2_160249478_160249680 | 0.33 |
Gm826 |
predicted gene 826 |
64037 |
0.13 |
| chr9_58719746_58719897 | 0.33 |
Rec114 |
REC114 meiotic recombination protein |
21738 |
0.18 |
| chr2_77516922_77517086 | 0.32 |
Zfp385b |
zinc finger protein 385B |
2531 |
0.37 |
| chr8_71378571_71378827 | 0.32 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
2210 |
0.15 |
| chr4_19527621_19527791 | 0.31 |
Cpne3 |
copine III |
15972 |
0.22 |
| chr16_38898912_38899113 | 0.31 |
Igsf11 |
immunoglobulin superfamily, member 11 |
2459 |
0.28 |
| chr17_34809326_34809625 | 0.31 |
C4a |
complement component 4A (Rodgers blood group) |
279 |
0.73 |
| chr11_98289682_98289855 | 0.31 |
Gm20644 |
predicted gene 20644 |
4386 |
0.11 |
| chr2_105854633_105854784 | 0.30 |
Elp4 |
elongator acetyltransferase complex subunit 4 |
11274 |
0.22 |
| chr3_27542592_27542743 | 0.30 |
Gm43344 |
predicted gene 43344 |
3974 |
0.29 |
| chr3_138289329_138289480 | 0.29 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
11753 |
0.11 |
| chr10_39624637_39624809 | 0.29 |
Gm16364 |
predicted gene 16364 |
11041 |
0.13 |
| chr8_94401407_94401558 | 0.29 |
Ap3s1-ps2 |
adaptor-related protein complex 3, sigma 1 subunit, pseudogene 2 |
3729 |
0.13 |
| chr12_110245003_110245222 | 0.29 |
4930425K24Rik |
RIKEN cDNA 4930425K24 gene |
8620 |
0.11 |
| chr16_93335454_93335612 | 0.29 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
17656 |
0.18 |
| chr4_100291152_100291321 | 0.29 |
Gm12706 |
predicted gene 12706 |
70823 |
0.12 |
| chr4_134372242_134372426 | 0.28 |
Extl1 |
exostosin-like glycosyltransferase 1 |
224 |
0.89 |
| chr7_17073907_17074066 | 0.28 |
Psg16 |
pregnancy specific glycoprotein 16 |
54 |
0.95 |
| chr8_34157897_34158198 | 0.27 |
Saraf |
store-operated calcium entry-associated regulatory factor |
434 |
0.71 |
| chr11_5912941_5913521 | 0.27 |
Gck |
glucokinase |
1893 |
0.2 |
| chr8_18619513_18619669 | 0.27 |
Gm25763 |
predicted gene, 25763 |
5482 |
0.18 |
| chr10_100131733_100131922 | 0.27 |
Gm22918 |
predicted gene, 22918 |
7674 |
0.14 |
| chr7_83637520_83637671 | 0.27 |
Gm45838 |
predicted gene 45838 |
4224 |
0.13 |
| chr2_83833142_83833332 | 0.26 |
Fam171b |
family with sequence similarity 171, member B |
20601 |
0.15 |
| chr11_53420114_53420567 | 0.26 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
2830 |
0.12 |
| chr15_80891104_80891265 | 0.25 |
Tnrc6b |
trinucleotide repeat containing 6b |
2034 |
0.31 |
| chr10_80963053_80963229 | 0.25 |
Gm3828 |
predicted gene 3828 |
8431 |
0.1 |
| chr4_58546190_58546357 | 0.25 |
Lpar1 |
lysophosphatidic acid receptor 1 |
2128 |
0.33 |
| chr10_18408613_18409116 | 0.24 |
Nhsl1 |
NHS-like 1 |
1189 |
0.56 |
| chr5_28042416_28042567 | 0.24 |
Gm43611 |
predicted gene 43611 |
6085 |
0.17 |
| chr2_164452146_164452320 | 0.24 |
Sys1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
4731 |
0.09 |
| chr6_94698690_94699533 | 0.24 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
1026 |
0.61 |
| chrX_142506860_142507020 | 0.24 |
Gm25915 |
predicted gene, 25915 |
21493 |
0.16 |
| chr6_91878620_91878777 | 0.24 |
Ccdc174 |
coiled-coil domain containing 174 |
623 |
0.45 |
| chr8_35462445_35462625 | 0.24 |
Eri1 |
exoribonuclease 1 |
18873 |
0.16 |
| chr6_3979259_3979410 | 0.24 |
Tfpi2 |
tissue factor pathway inhibitor 2 |
9524 |
0.17 |
| chr16_42911745_42911939 | 0.24 |
Gm19522 |
predicted gene, 19522 |
223 |
0.93 |
| chr12_85487294_85487804 | 0.24 |
Fos |
FBJ osteosarcoma oncogene |
12638 |
0.16 |
| chr17_27027927_27028127 | 0.23 |
Bak1 |
BCL2-antagonist/killer 1 |
551 |
0.52 |
| chr4_70044084_70044244 | 0.23 |
Gm11225 |
predicted gene 11225 |
2993 |
0.36 |
| chr13_69631437_69631671 | 0.23 |
Nsun2 |
NOL1/NOP2/Sun domain family member 2 |
814 |
0.46 |
| chr4_125419601_125419754 | 0.23 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
71023 |
0.1 |
| chr13_44420237_44420395 | 0.23 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
19396 |
0.14 |
| chr7_128612566_128612733 | 0.23 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
1289 |
0.33 |
| chr9_105053654_105053819 | 0.23 |
Mrpl3 |
mitochondrial ribosomal protein L3 |
435 |
0.77 |
| chr17_10235141_10235292 | 0.23 |
Qk |
quaking |
21642 |
0.25 |
| chr5_150578974_150579265 | 0.23 |
N4bp2l1 |
NEDD4 binding protein 2-like 1 |
2128 |
0.14 |
| chr8_41216648_41216810 | 0.23 |
Gm16348 |
predicted gene 16348 |
1410 |
0.33 |
| chr1_91903121_91903295 | 0.22 |
Gm37600 |
predicted gene, 37600 |
13052 |
0.19 |
| chr10_81419362_81419546 | 0.22 |
Mir1191b |
microRNA 1191b |
3157 |
0.08 |
| chr11_97776781_97777120 | 0.22 |
1700001P01Rik |
RIKEN cDNA 1700001P01 gene |
1032 |
0.25 |
| chr10_75409660_75410057 | 0.22 |
Upb1 |
ureidopropionase, beta |
185 |
0.94 |
| chr15_89084694_89084845 | 0.22 |
Trabd |
TraB domain containing |
60 |
0.94 |
| chr5_66085295_66085580 | 0.22 |
Gm43323 |
predicted gene 43323 |
971 |
0.43 |
| chr12_59012425_59012608 | 0.22 |
Sec23a |
SEC23 homolog A, COPII coat complex component |
499 |
0.58 |
| chr4_123990670_123991070 | 0.22 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr4_55231638_55231798 | 0.22 |
Gm12508 |
predicted gene 12508 |
23022 |
0.15 |
| chr7_97424594_97424745 | 0.22 |
Thrsp |
thyroid hormone responsive |
6939 |
0.13 |
| chr13_47104820_47104971 | 0.22 |
Dek |
DEK oncogene (DNA binding) |
935 |
0.39 |
| chr8_36252100_36252286 | 0.21 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
2677 |
0.29 |
| chr3_53863181_53863486 | 0.21 |
Ufm1 |
ubiquitin-fold modifier 1 |
356 |
0.86 |
| chr8_48980844_48981032 | 0.21 |
Gm25830 |
predicted gene, 25830 |
22457 |
0.22 |
| chr19_32769635_32769845 | 0.21 |
Pten |
phosphatase and tensin homolog |
12145 |
0.23 |
| chr4_120074199_120074385 | 0.20 |
AL607142.1 |
novel protein |
9296 |
0.22 |
| chr8_45794944_45795095 | 0.20 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
8189 |
0.16 |
| chr18_65803330_65803542 | 0.20 |
Sec11c |
SEC11 homolog C, signal peptidase complex subunit |
2134 |
0.21 |
| chr15_68260157_68260308 | 0.20 |
Zfat |
zinc finger and AT hook domain containing |
1376 |
0.43 |
| chr1_127222412_127222573 | 0.20 |
Mgat5 |
mannoside acetylglucosaminyltransferase 5 |
17464 |
0.17 |
| chr5_90891820_90892079 | 0.20 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
706 |
0.49 |
| chr6_82085852_82086023 | 0.20 |
Gm15864 |
predicted gene 15864 |
33356 |
0.15 |
| chr5_9042843_9042994 | 0.20 |
Gm40264 |
predicted gene, 40264 |
7794 |
0.15 |
| chr6_144170659_144171019 | 0.20 |
Sox5 |
SRY (sex determining region Y)-box 5 |
33218 |
0.23 |
| chr2_35102321_35102587 | 0.20 |
AI182371 |
expressed sequence AI182371 |
911 |
0.53 |
| chr9_50835575_50835937 | 0.20 |
Alg9 |
asparagine-linked glycosylation 9 (alpha 1,2 mannosyltransferase) |
185 |
0.93 |
| chr10_44529721_44529883 | 0.20 |
Prdm1 |
PR domain containing 1, with ZNF domain |
1301 |
0.42 |
| chr6_5634324_5634496 | 0.19 |
Dync1i1 |
dynein cytoplasmic 1 intermediate chain 1 |
91229 |
0.09 |
| chr1_136662300_136662465 | 0.19 |
Gm19705 |
predicted gene, 19705 |
8387 |
0.13 |
| chr19_21663399_21663699 | 0.19 |
Abhd17b |
abhydrolase domain containing 17B |
10024 |
0.2 |
| chr4_109116360_109116524 | 0.19 |
Osbpl9 |
oxysterol binding protein-like 9 |
1256 |
0.5 |
| chr14_41150467_41150746 | 0.19 |
Mbl1 |
mannose-binding lectin (protein A) 1 |
852 |
0.42 |
| chr2_148112041_148112203 | 0.19 |
Gm24221 |
predicted gene, 24221 |
39028 |
0.13 |
| chr17_28693242_28693406 | 0.19 |
Mapk14 |
mitogen-activated protein kinase 14 |
756 |
0.53 |
| chr18_45563342_45563666 | 0.19 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
3350 |
0.27 |
| chr4_147452443_147452608 | 0.19 |
Vmn2r-ps19 |
vomeronasal 2, receptor, pseudogene 19 |
8145 |
0.13 |
| chr5_147548677_147549145 | 0.19 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
9634 |
0.2 |
| chr14_34663553_34663710 | 0.19 |
Gm49024 |
predicted gene, 49024 |
1924 |
0.23 |
| chr9_43746925_43747076 | 0.19 |
Gm30015 |
predicted gene, 30015 |
900 |
0.49 |
| chr8_70216821_70217037 | 0.19 |
Slc25a42 |
solute carrier family 25, member 42 |
4624 |
0.11 |
| chr17_26602213_26602380 | 0.19 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
72 |
0.96 |
| chr11_60538899_60539087 | 0.19 |
Alkbh5 |
alkB homolog 5, RNA demethylase |
1015 |
0.34 |
| chr17_46735081_46735246 | 0.18 |
Cnpy3 |
canopy FGF signaling regulator 3 |
2464 |
0.14 |
| chr13_51902467_51902673 | 0.18 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
55826 |
0.12 |
| chr7_24573916_24574086 | 0.18 |
Xrcc1 |
X-ray repair complementing defective repair in Chinese hamster cells 1 |
1775 |
0.17 |
| chr9_106261489_106261640 | 0.18 |
Alas1 |
aminolevulinic acid synthase 1 |
12910 |
0.1 |
| chr12_21142640_21142894 | 0.18 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30813 |
0.16 |
| chr13_4435934_4436251 | 0.18 |
Akr1c6 |
aldo-keto reductase family 1, member C6 |
54 |
0.97 |
| chr1_21230019_21230187 | 0.18 |
Gm38224 |
predicted gene, 38224 |
4619 |
0.13 |
| chr19_44397401_44397552 | 0.17 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
9214 |
0.14 |
| chr4_126119139_126119304 | 0.17 |
Stk40 |
serine/threonine kinase 40 |
949 |
0.41 |
| chr8_88023561_88023721 | 0.17 |
Gm2716 |
predicted gene 2716 |
47169 |
0.12 |
| chr11_120683455_120683634 | 0.17 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
3149 |
0.08 |
| chr5_93026523_93026714 | 0.17 |
Gm25521 |
predicted gene, 25521 |
13479 |
0.13 |
| chr11_70741997_70742156 | 0.17 |
Gm12320 |
predicted gene 12320 |
13075 |
0.08 |
| chr10_81545615_81545793 | 0.17 |
Gna11 |
guanine nucleotide binding protein, alpha 11 |
514 |
0.55 |
| chr6_140196798_140197143 | 0.17 |
Gm24174 |
predicted gene, 24174 |
30325 |
0.16 |
| chr10_24835899_24836097 | 0.17 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
115 |
0.96 |
| chr9_106160482_106160686 | 0.17 |
Glyctk |
glycerate kinase |
2446 |
0.13 |
| chr4_104986841_104987054 | 0.17 |
Gm12721 |
predicted gene 12721 |
16818 |
0.24 |
| chr18_53868371_53868522 | 0.17 |
Csnk1g3 |
casein kinase 1, gamma 3 |
6258 |
0.3 |
| chr10_29311597_29311753 | 0.17 |
Echdc1 |
enoyl Coenzyme A hydratase domain containing 1 |
1491 |
0.26 |
| chr15_59278504_59278870 | 0.17 |
Gm36617 |
predicted gene, 36617 |
3291 |
0.21 |
| chr11_52053703_52053862 | 0.17 |
Gm12205 |
predicted gene 12205 |
14311 |
0.12 |
| chr4_106777883_106778393 | 0.17 |
Gm12745 |
predicted gene 12745 |
3322 |
0.19 |
| chr4_123016748_123017047 | 0.17 |
Trit1 |
tRNA isopentenyltransferase 1 |
300 |
0.87 |
| chrX_72918774_72918925 | 0.16 |
Nsdhl |
NAD(P) dependent steroid dehydrogenase-like |
292 |
0.61 |
| chr7_99119375_99119533 | 0.16 |
Gm44975 |
predicted gene 44975 |
4005 |
0.16 |
| chr17_83843855_83844136 | 0.16 |
Haao |
3-hydroxyanthranilate 3,4-dioxygenase |
2754 |
0.24 |
| chr18_44540954_44541143 | 0.16 |
Mcc |
mutated in colorectal cancers |
21532 |
0.24 |
| chr12_71904296_71904706 | 0.16 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
14771 |
0.22 |
| chr12_13730330_13730517 | 0.16 |
Gm4929 |
predicted gene 4929 |
23242 |
0.16 |
| chr6_87804742_87804969 | 0.16 |
9930120I10Rik |
RIKEN cDNA 9930120I10 gene |
1481 |
0.18 |
| chr3_31133502_31133712 | 0.16 |
Cldn11 |
claudin 11 |
16313 |
0.19 |
| chr6_29712151_29712310 | 0.16 |
Tspan33 |
tetraspanin 33 |
4202 |
0.19 |
| chr18_11658016_11658319 | 0.16 |
Rbbp8 |
retinoblastoma binding protein 8, endonuclease |
325 |
0.54 |
| chr16_50732953_50733143 | 0.16 |
Dubr |
Dppa2 upstream binding RNA |
275 |
0.91 |
| chr7_98351056_98351207 | 0.15 |
Tsku |
tsukushi, small leucine rich proteoglycan |
8948 |
0.17 |
| chr2_48981052_48981464 | 0.15 |
Orc4 |
origin recognition complex, subunit 4 |
30981 |
0.18 |
| chr18_64641731_64641886 | 0.15 |
Atp8b1 |
ATPase, class I, type 8B, member 1 |
18999 |
0.13 |
| chr19_47315824_47316178 | 0.15 |
Sh3pxd2a |
SH3 and PX domains 2A |
1250 |
0.45 |
| chr8_24160713_24160865 | 0.15 |
A730045E13Rik |
RIKEN cDNA A730045E13 gene |
13115 |
0.25 |
| chr9_57312626_57312793 | 0.15 |
Gm18996 |
predicted gene, 18996 |
16706 |
0.14 |
| chr8_119440803_119440970 | 0.15 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
3697 |
0.18 |
| chr7_13054073_13054266 | 0.15 |
Mzf1 |
myeloid zinc finger 1 |
172 |
0.88 |
| chr9_46481072_46481223 | 0.15 |
Gm47144 |
predicted gene, 47144 |
23926 |
0.16 |
| chr6_48676261_48676422 | 0.15 |
Gimap9 |
GTPase, IMAP family member 9 |
212 |
0.83 |
| chr19_53311508_53311850 | 0.14 |
Mxi1 |
MAX interactor 1, dimerization protein |
1161 |
0.41 |
| chr6_144155774_144155925 | 0.14 |
Sox5 |
SRY (sex determining region Y)-box 5 |
48208 |
0.18 |
| chr3_10290182_10290333 | 0.14 |
Fabp12 |
fatty acid binding protein 12 |
10917 |
0.1 |
| chr14_70712939_70713321 | 0.14 |
Xpo7 |
exportin 7 |
5095 |
0.18 |
| chr13_23729407_23729642 | 0.14 |
Gm11338 |
predicted gene 11338 |
976 |
0.2 |
| chr14_65136751_65136947 | 0.14 |
Extl3 |
exostosin-like glycosyltransferase 3 |
13006 |
0.15 |
| chr4_125127317_125127672 | 0.14 |
Zc3h12a |
zinc finger CCCH type containing 12A |
346 |
0.85 |
| chr5_73360537_73360690 | 0.14 |
Ociad2 |
OCIA domain containing 2 |
19585 |
0.1 |
| chr4_101250022_101250385 | 0.14 |
Jak1 |
Janus kinase 1 |
15020 |
0.13 |
| chr11_60827336_60827487 | 0.14 |
Dhrs7b |
dehydrogenase/reductase (SDR family) member 7B |
3220 |
0.12 |
| chr18_67961364_67961594 | 0.14 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
28222 |
0.18 |
| chr10_127414051_127414202 | 0.14 |
R3hdm2 |
R3H domain containing 2 |
6212 |
0.12 |
| chr1_59387773_59387951 | 0.14 |
Gm29016 |
predicted gene 29016 |
8932 |
0.2 |
| chr2_18059830_18059981 | 0.14 |
Mir7655 |
microRNA 7655 |
2059 |
0.18 |
| chrX_98051224_98051403 | 0.14 |
Gm14802 |
predicted gene 14802 |
10938 |
0.3 |
| chr2_25210906_25211266 | 0.14 |
Nelfb |
negative elongation factor complex member B |
295 |
0.68 |
| chr1_21262098_21262745 | 0.14 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8900 |
0.11 |
| chr8_122576204_122576481 | 0.14 |
Aprt |
adenine phosphoribosyl transferase |
228 |
0.84 |
| chr1_71999089_71999255 | 0.14 |
Gm5528 |
predicted gene 5528 |
5116 |
0.18 |
| chr11_106841639_106841811 | 0.14 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
2427 |
0.19 |
| chr1_87891961_87892159 | 0.13 |
Gm38365 |
predicted gene, 38365 |
5480 |
0.13 |
| chr13_47171335_47171540 | 0.13 |
Gm48581 |
predicted gene, 48581 |
16429 |
0.14 |
| chr9_121118120_121118389 | 0.13 |
Ulk4 |
unc-51-like kinase 4 |
9729 |
0.23 |
| chr3_93600843_93601099 | 0.13 |
Gm5541 |
predicted gene 5541 |
28781 |
0.09 |
| chr5_140112771_140112922 | 0.13 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
2620 |
0.23 |
| chr12_87348210_87348371 | 0.13 |
Sptlc2 |
serine palmitoyltransferase, long chain base subunit 2 |
2091 |
0.19 |
| chr18_46607931_46608095 | 0.13 |
Eif1a |
eukaryotic translation initiation factor 1A |
9999 |
0.14 |
| chr18_76930180_76930397 | 0.13 |
Gm50364 |
predicted gene, 50364 |
162 |
0.58 |
| chr6_115717866_115718504 | 0.13 |
Gm24008 |
predicted gene, 24008 |
5647 |
0.12 |
| chr10_115314574_115315134 | 0.13 |
Rab21 |
RAB21, member RAS oncogene family |
737 |
0.41 |
| chr4_49435071_49435222 | 0.13 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
15988 |
0.12 |
| chr3_89864780_89864960 | 0.13 |
She |
src homology 2 domain-containing transforming protein E |
26981 |
0.1 |
| chr4_59242009_59242160 | 0.13 |
Gm12596 |
predicted gene 12596 |
17967 |
0.17 |
| chr6_57523099_57523250 | 0.13 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
7567 |
0.15 |
| chr6_128526251_128526407 | 0.13 |
Pzp |
PZP, alpha-2-macroglobulin like |
374 |
0.73 |
| chr19_40512528_40512992 | 0.13 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
913 |
0.57 |
| chr10_97060750_97060908 | 0.13 |
4930556N09Rik |
RIKEN cDNA 4930556N09 gene |
25049 |
0.2 |
| chr6_94193888_94194039 | 0.13 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
89062 |
0.09 |
| chr2_126597028_126597385 | 0.13 |
Hdc |
histidine decarboxylase |
1055 |
0.5 |
| chr4_45505286_45505685 | 0.13 |
Gm22518 |
predicted gene, 22518 |
4933 |
0.17 |
| chr13_56134831_56134982 | 0.13 |
Macroh2a1 |
macroH2A.1 histone |
81 |
0.97 |
| chr2_32601599_32601765 | 0.13 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
1956 |
0.14 |
| chr1_74047431_74047619 | 0.13 |
Tns1 |
tensin 1 |
10368 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.1 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |