Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
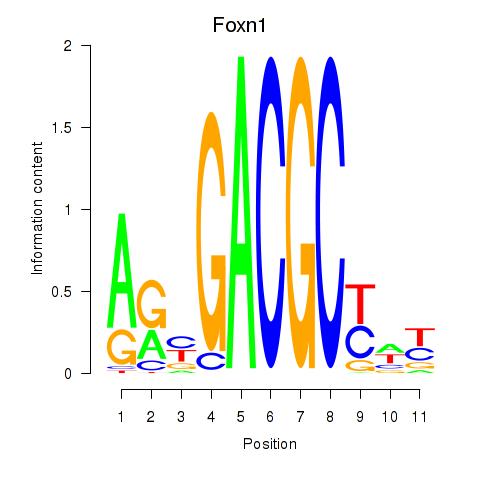
Results for Foxn1
Z-value: 1.05
Transcription factors associated with Foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxn1
|
ENSMUSG00000002057.4 | forkhead box N1 |
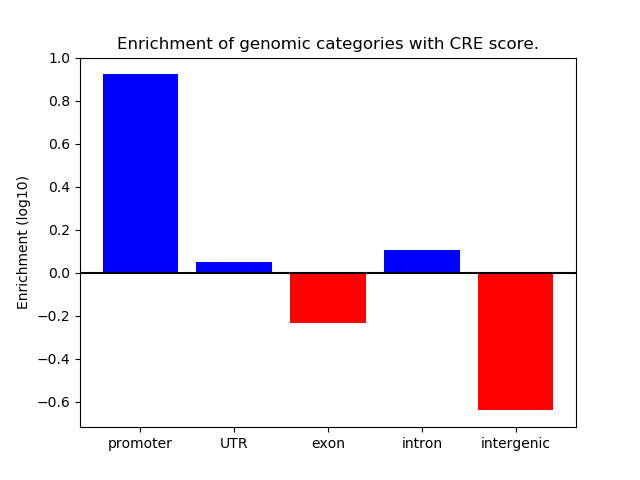
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_78372687_78372853 | Foxn1 | 13788 | 0.087005 | 0.26 | 6.1e-01 | Click! |
| chr11_78372975_78373151 | Foxn1 | 13495 | 0.087385 | 0.22 | 6.8e-01 | Click! |
Activity of the Foxn1 motif across conditions
Conditions sorted by the z-value of the Foxn1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
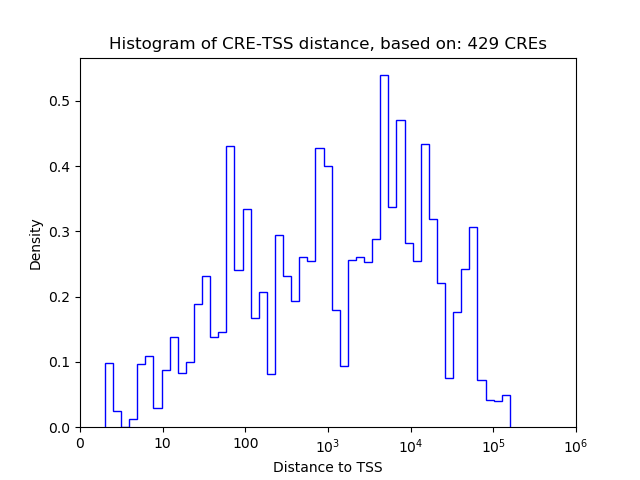
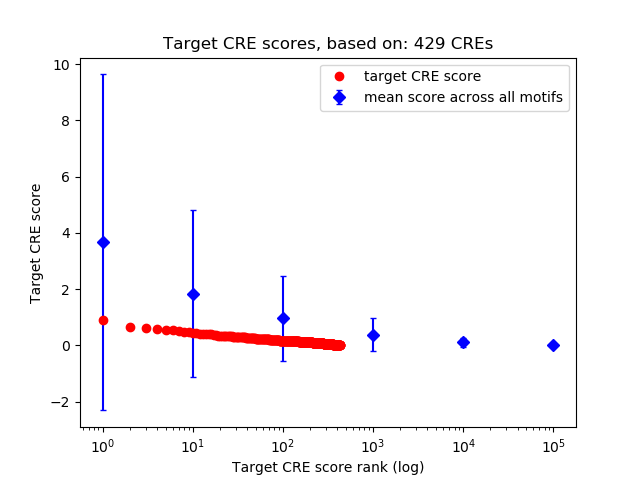
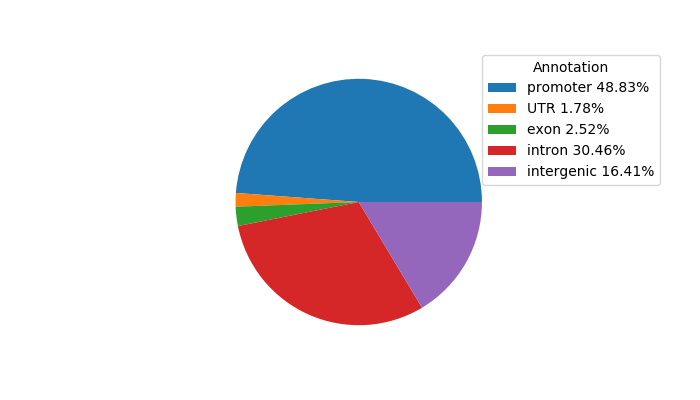
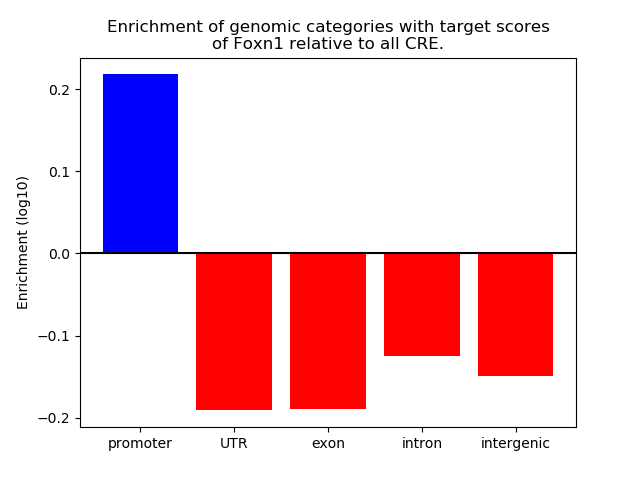
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_51089937_51090106 | 0.89 |
Rnase4 |
ribonuclease, RNase A family 4 |
1056 |
0.24 |
| chr8_11249894_11250051 | 0.64 |
Col4a1 |
collagen, type IV, alpha 1 |
4367 |
0.21 |
| chr7_48930338_48930500 | 0.62 |
Gm18559 |
predicted gene, 18559 |
9699 |
0.14 |
| chr2_72179596_72179778 | 0.57 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
6 |
0.98 |
| chr14_52279190_52279559 | 0.56 |
Rab2b |
RAB2B, member RAS oncogene family |
35 |
0.68 |
| chr5_113142564_113142931 | 0.53 |
4930557B06Rik |
RIKEN cDNA 4930557B06 gene |
298 |
0.81 |
| chr10_40302183_40302345 | 0.53 |
Amd1 |
S-adenosylmethionine decarboxylase 1 |
76 |
0.95 |
| chr9_43636478_43636893 | 0.49 |
Gm29961 |
predicted gene, 29961 |
7393 |
0.18 |
| chr1_171281170_171281333 | 0.47 |
Ppox |
protoporphyrinogen oxidase |
65 |
0.91 |
| chr9_63201496_63201967 | 0.44 |
Skor1 |
SKI family transcriptional corepressor 1 |
52770 |
0.12 |
| chr2_148522150_148522301 | 0.43 |
Gm14120 |
predicted gene 14120 |
53335 |
0.11 |
| chr1_157008781_157008932 | 0.41 |
Gm10531 |
predicted gene 10531 |
34691 |
0.13 |
| chr9_104161174_104161357 | 0.40 |
Dnajc13 |
DnaJ heat shock protein family (Hsp40) member C13 |
656 |
0.61 |
| chr1_180161597_180161764 | 0.40 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
673 |
0.64 |
| chr6_128803059_128803375 | 0.40 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
14002 |
0.09 |
| chr11_109670186_109670371 | 0.40 |
Prkar1a |
protein kinase, cAMP dependent regulatory, type I, alpha |
4334 |
0.19 |
| chr19_27429919_27430086 | 0.38 |
Pum3 |
pumilio RNA-binding family member 3 |
177 |
0.96 |
| chr2_110229824_110230205 | 0.35 |
Gm22997 |
predicted gene, 22997 |
12011 |
0.18 |
| chr17_30609874_30610226 | 0.35 |
Glo1 |
glyoxalase 1 |
2519 |
0.15 |
| chr8_121071062_121071213 | 0.34 |
Fendrr |
Foxf1 adjacent non-coding developmental regulatory RNA |
10724 |
0.12 |
| chr15_54924268_54924437 | 0.34 |
Enpp2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
1739 |
0.36 |
| chr13_113589653_113589822 | 0.34 |
Gm34471 |
predicted gene, 34471 |
21970 |
0.15 |
| chr13_54622139_54622505 | 0.33 |
Faf2 |
Fas associated factor family member 2 |
484 |
0.69 |
| chr3_104639553_104639867 | 0.33 |
Slc16a1 |
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
1042 |
0.32 |
| chr13_40890430_40890581 | 0.32 |
Gcnt2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
3312 |
0.14 |
| chr14_54603440_54603604 | 0.32 |
4931414P19Rik |
RIKEN cDNA 4931414P19 gene |
2471 |
0.12 |
| chr10_13553038_13553200 | 0.32 |
Pex3 |
peroxisomal biogenesis factor 3 |
1 |
0.75 |
| chr19_6242419_6242590 | 0.31 |
Atg2a |
autophagy related 2A |
836 |
0.36 |
| chr2_29124935_29125101 | 0.31 |
Setx |
senataxin |
26 |
0.97 |
| chr16_30965341_30965552 | 0.30 |
Gm15742 |
predicted gene 15742 |
8072 |
0.18 |
| chrX_12134649_12134805 | 0.30 |
Bcor |
BCL6 interacting corepressor |
6327 |
0.27 |
| chr10_18065992_18066156 | 0.30 |
Reps1 |
RalBP1 associated Eps domain containing protein |
10070 |
0.2 |
| chr7_35396851_35397006 | 0.30 |
Faap24 |
Fanconi anemia core complex associated protein 24 |
92 |
0.51 |
| chr4_132040365_132040540 | 0.29 |
Gm13063 |
predicted gene 13063 |
7726 |
0.12 |
| chr1_93123310_93123471 | 0.29 |
Gm28086 |
predicted gene 28086 |
713 |
0.57 |
| chr10_99576469_99576639 | 0.29 |
Gm20110 |
predicted gene, 20110 |
32617 |
0.14 |
| chr2_19658586_19658737 | 0.28 |
Otud1 |
OTU domain containing 1 |
909 |
0.47 |
| chr7_28831672_28831952 | 0.28 |
Ech1 |
enoyl coenzyme A hydratase 1, peroxisomal |
561 |
0.52 |
| chr13_14063374_14063726 | 0.27 |
Ggps1 |
geranylgeranyl diphosphate synthase 1 |
62 |
0.72 |
| chr7_80088340_80088502 | 0.27 |
Gm44949 |
predicted gene 44949 |
2010 |
0.2 |
| chr11_100738407_100738714 | 0.27 |
Rab5c |
RAB5C, member RAS oncogene family |
345 |
0.76 |
| chr9_24420197_24420363 | 0.27 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
3679 |
0.27 |
| chr3_89418996_89419160 | 0.26 |
Shc1 |
src homology 2 domain-containing transforming protein C1 |
464 |
0.47 |
| chr1_75253990_75254151 | 0.26 |
Gm28294 |
predicted gene 28294 |
1748 |
0.15 |
| chr11_98982083_98982254 | 0.26 |
Gjd3 |
gap junction protein, delta 3 |
848 |
0.44 |
| chr8_22335552_22335714 | 0.26 |
Tpte |
transmembrane phosphatase with tensin homology |
52192 |
0.08 |
| chr9_29750692_29750843 | 0.26 |
Gm15521 |
predicted gene 15521 |
158257 |
0.04 |
| chr12_25235200_25235794 | 0.26 |
Gm19340 |
predicted gene, 19340 |
47098 |
0.12 |
| chr8_126527053_126527635 | 0.25 |
Gm26759 |
predicted gene, 26759 |
39626 |
0.14 |
| chr1_23187785_23188477 | 0.24 |
Gm29506 |
predicted gene 29506 |
47542 |
0.11 |
| chr13_58067763_58068069 | 0.24 |
Klhl3 |
kelch-like 3 |
34512 |
0.13 |
| chr5_53992615_53992786 | 0.24 |
Stim2 |
stromal interaction molecule 2 |
5799 |
0.26 |
| chr6_134792589_134792746 | 0.23 |
Dusp16 |
dual specificity phosphatase 16 |
42 |
0.97 |
| chr5_117417582_117417750 | 0.23 |
Ksr2 |
kinase suppressor of ras 2 |
3666 |
0.18 |
| chr4_107002389_107002540 | 0.23 |
Gm12786 |
predicted gene 12786 |
20035 |
0.15 |
| chr15_97019476_97019938 | 0.23 |
Slc38a4 |
solute carrier family 38, member 4 |
244 |
0.95 |
| chr1_193288543_193288694 | 0.23 |
Gm37650 |
predicted gene, 37650 |
2271 |
0.18 |
| chr12_102748209_102748378 | 0.23 |
Tmem251 |
transmembrane protein 251 |
4531 |
0.09 |
| chr19_38089508_38089672 | 0.23 |
Ffar4 |
free fatty acid receptor 4 |
7481 |
0.14 |
| chr7_80653838_80654052 | 0.23 |
Gm15880 |
predicted gene 15880 |
17928 |
0.15 |
| chr11_120340498_120340661 | 0.22 |
Gm23419 |
predicted gene, 23419 |
6240 |
0.09 |
| chr10_83347610_83347786 | 0.22 |
Gm40723 |
predicted gene, 40723 |
3185 |
0.21 |
| chr5_130171601_130171776 | 0.22 |
Rabgef1 |
RAB guanine nucleotide exchange factor (GEF) 1 |
110 |
0.94 |
| chr8_119435449_119435786 | 0.22 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
1493 |
0.34 |
| chr10_17722281_17722461 | 0.22 |
Cited2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
847 |
0.66 |
| chr18_68499103_68499266 | 0.21 |
Gm29677 |
predicted gene, 29677 |
60238 |
0.1 |
| chr11_6335474_6335637 | 0.21 |
Ogdh |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
42950 |
0.08 |
| chr10_81551987_81552144 | 0.21 |
Gna11 |
guanine nucleotide binding protein, alpha 11 |
6875 |
0.07 |
| chr9_37147174_37147330 | 0.21 |
Pknox2 |
Pbx/knotted 1 homeobox 2 |
14 |
0.96 |
| chr9_124424931_124425085 | 0.21 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
854 |
0.55 |
| chr11_100808840_100809039 | 0.20 |
Stat5b |
signal transducer and activator of transcription 5B |
13589 |
0.12 |
| chr7_28437107_28437270 | 0.20 |
Gmfg |
glia maturation factor, gamma |
259 |
0.76 |
| chr4_132946310_132946569 | 0.20 |
Gm24913 |
predicted gene, 24913 |
15122 |
0.13 |
| chr8_66486485_66486874 | 0.20 |
Tma16 |
translation machinery associated 16 |
149 |
0.95 |
| chr10_66909327_66909486 | 0.20 |
1110002J07Rik |
RIKEN cDNA 1110002J07 gene |
8333 |
0.16 |
| chr1_74124750_74124964 | 0.19 |
Tns1 |
tensin 1 |
408 |
0.62 |
| chr14_79482321_79482598 | 0.19 |
Wbp4 |
WW domain binding protein 4 |
939 |
0.41 |
| chr2_94162272_94162444 | 0.19 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
4394 |
0.19 |
| chr13_14064834_14065347 | 0.19 |
Arid4b |
AT rich interactive domain 4B (RBP1-like) |
267 |
0.84 |
| chr6_93876489_93876704 | 0.19 |
Gm22840 |
predicted gene, 22840 |
14019 |
0.23 |
| chr15_31344240_31344565 | 0.18 |
Ankrd33b |
ankyrin repeat domain 33B |
19153 |
0.14 |
| chr4_84069800_84069979 | 0.18 |
6030471H07Rik |
RIKEN cDNA 6030471H07 gene |
25010 |
0.19 |
| chr4_152302091_152302261 | 0.18 |
Icmt |
isoprenylcysteine carboxyl methyltransferase |
3211 |
0.14 |
| chr11_98740713_98740882 | 0.18 |
Thra |
thyroid hormone receptor alpha |
4 |
0.95 |
| chr19_41388446_41388602 | 0.18 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
3422 |
0.3 |
| chr7_12950233_12950384 | 0.18 |
1810019N24Rik |
RIKEN cDNA 1810019N24 gene |
242 |
0.81 |
| chr9_96758999_96759348 | 0.17 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
6342 |
0.16 |
| chr17_29323723_29323898 | 0.17 |
Gm46603 |
predicted gene, 46603 |
1261 |
0.28 |
| chr3_105825808_105826159 | 0.17 |
Gm5547 |
predicted gene 5547 |
9370 |
0.12 |
| chr13_52828377_52828538 | 0.17 |
BB123696 |
expressed sequence BB123696 |
71252 |
0.11 |
| chr14_114854717_114854875 | 0.17 |
Gm49010 |
predicted gene, 49010 |
21714 |
0.15 |
| chr3_105053004_105053165 | 0.17 |
Cttnbp2nl |
CTTNBP2 N-terminal like |
62 |
0.97 |
| chr3_95928829_95929106 | 0.17 |
Anp32e |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
279 |
0.81 |
| chr10_117497904_117498070 | 0.17 |
Gm32141 |
predicted gene, 32141 |
50494 |
0.09 |
| chr14_62556439_62556816 | 0.17 |
Fam124a |
family with sequence similarity 124, member A |
852 |
0.48 |
| chr17_26123354_26123516 | 0.17 |
Mrpl28 |
mitochondrial ribosomal protein L28 |
65 |
0.93 |
| chr9_58582606_58582757 | 0.16 |
Nptn |
neuroplastin |
237 |
0.92 |
| chr8_11173356_11173731 | 0.16 |
Gm15418 |
predicted gene 15418 |
14212 |
0.15 |
| chr2_132110806_132111002 | 0.16 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
544 |
0.75 |
| chr16_55698500_55698846 | 0.16 |
Gm19771 |
predicted gene, 19771 |
41245 |
0.16 |
| chr3_79218581_79218770 | 0.16 |
4921511C10Rik |
RIKEN cDNA 4921511C10 gene |
5756 |
0.2 |
| chr15_100121547_100121719 | 0.16 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
13472 |
0.14 |
| chr4_132515539_132515706 | 0.16 |
Sesn2 |
sestrin 2 |
5121 |
0.1 |
| chr19_47438467_47438632 | 0.16 |
Sh3pxd2a |
SH3 and PX domains 2A |
25698 |
0.17 |
| chr17_35918919_35919191 | 0.16 |
Mir1894 |
microRNA 1894 |
1166 |
0.17 |
| chr1_57429882_57430054 | 0.16 |
Gm22371 |
predicted gene, 22371 |
7595 |
0.15 |
| chr5_96917597_96917811 | 0.16 |
Gm8013 |
predicted gene 8013 |
3568 |
0.12 |
| chr2_131222260_131222415 | 0.16 |
Mavs |
mitochondrial antiviral signaling protein |
11726 |
0.1 |
| chr1_72288908_72289079 | 0.16 |
Tmem169 |
transmembrane protein 169 |
4598 |
0.15 |
| chr12_80247162_80247339 | 0.16 |
Actn1 |
actinin, alpha 1 |
11534 |
0.15 |
| chr11_31671872_31672045 | 0.16 |
Bod1 |
biorientation of chromosomes in cell division 1 |
73 |
0.98 |
| chr10_79936936_79937107 | 0.16 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
6597 |
0.06 |
| chr7_70549056_70549216 | 0.16 |
Gm35842 |
predicted gene, 35842 |
267 |
0.88 |
| chr7_141581336_141581865 | 0.16 |
Ap2a2 |
adaptor-related protein complex 2, alpha 2 subunit |
19070 |
0.1 |
| chr10_8844879_8845170 | 0.16 |
4930553I21Rik |
RIKEN cDNA 4930553I21 gene |
16419 |
0.15 |
| chr3_146450379_146450546 | 0.16 |
Ctbs |
chitobiase |
8 |
0.97 |
| chr17_28930498_28930665 | 0.15 |
Gm16191 |
predicted gene 16191 |
3840 |
0.1 |
| chr16_86816748_86816918 | 0.15 |
Gm32624 |
predicted gene, 32624 |
33 |
0.98 |
| chr8_106278256_106278407 | 0.15 |
Smpd3 |
sphingomyelin phosphodiesterase 3, neutral |
2647 |
0.22 |
| chr11_54539316_54539467 | 0.15 |
Rapgef6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
16445 |
0.19 |
| chr7_24972671_24972836 | 0.15 |
Rabac1 |
Rab acceptor 1 (prenylated) |
1 |
0.95 |
| chr9_69273022_69273200 | 0.15 |
Rora |
RAR-related orphan receptor alpha |
16571 |
0.24 |
| chr1_55088026_55088196 | 0.15 |
Hspe1 |
heat shock protein 1 (chaperonin 10) |
21 |
0.55 |
| chr13_81783090_81783247 | 0.15 |
Cetn3 |
centrin 3 |
91 |
0.9 |
| chr15_27919308_27919503 | 0.15 |
Trio |
triple functional domain (PTPRF interacting) |
134 |
0.97 |
| chr15_90215048_90215209 | 0.15 |
Alg10b |
asparagine-linked glycosylation 10B (alpha-1,2-glucosyltransferase) |
9183 |
0.22 |
| chr9_70207215_70207378 | 0.15 |
Myo1e |
myosin IE |
54 |
0.98 |
| chr14_46615984_46616316 | 0.15 |
Gm49319 |
predicted gene, 49319 |
5140 |
0.13 |
| chr5_145191503_145191838 | 0.15 |
Atp5j2 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
78 |
0.94 |
| chr16_97357218_97357390 | 0.15 |
Bace2 |
beta-site APP-cleaving enzyme 2 |
562 |
0.8 |
| chr9_99568726_99568886 | 0.15 |
Armc8 |
armadillo repeat containing 8 |
93 |
0.96 |
| chr15_95897769_95897945 | 0.15 |
Gm25070 |
predicted gene, 25070 |
18040 |
0.17 |
| chr10_93523037_93523409 | 0.14 |
Amdhd1 |
amidohydrolase domain containing 1 |
16810 |
0.12 |
| chr4_123718182_123718341 | 0.14 |
Ndufs5 |
NADH:ubiquinone oxidoreductase core subunit S5 |
59 |
0.96 |
| chr19_8920279_8920446 | 0.14 |
B3gat3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
12 |
0.91 |
| chr11_116103660_116103816 | 0.14 |
Trim47 |
tripartite motif-containing 47 |
3345 |
0.12 |
| chrX_7841056_7841418 | 0.14 |
Otud5 |
OTU domain containing 5 |
127 |
0.91 |
| chr14_65033978_65034138 | 0.14 |
Extl3 |
exostosin-like glycosyltransferase 3 |
64047 |
0.09 |
| chr16_14158957_14159171 | 0.14 |
Marf1 |
meiosis regulator and mRNA stability 1 |
203 |
0.78 |
| chr18_65490248_65490399 | 0.14 |
Gm50246 |
predicted gene, 50246 |
11083 |
0.13 |
| chr9_110871597_110871767 | 0.14 |
Tmie |
transmembrane inner ear |
4339 |
0.1 |
| chr9_63611147_63611298 | 0.14 |
Aagab |
alpha- and gamma-adaptin binding protein |
5766 |
0.2 |
| chr18_9957685_9957884 | 0.14 |
Thoc1 |
THO complex 1 |
122 |
0.96 |
| chr8_106936076_106936231 | 0.14 |
Sntb2 |
syntrophin, basic 2 |
4 |
0.96 |
| chr6_85451857_85452015 | 0.14 |
Pradc1 |
protease-associated domain containing 1 |
9 |
0.55 |
| chr16_26105727_26105925 | 0.14 |
P3h2 |
prolyl 3-hydroxylase 2 |
42 |
0.99 |
| chr6_54593211_54593386 | 0.14 |
Fkbp14 |
FK506 binding protein 14 |
121 |
0.95 |
| chr11_69671384_69671748 | 0.14 |
Eif4a1 |
eukaryotic translation initiation factor 4A1 |
116 |
0.83 |
| chr6_113483178_113483358 | 0.14 |
Creld1 |
cysteine-rich with EGF-like domains 1 |
29 |
0.94 |
| chr2_135711190_135711341 | 0.14 |
Gm14211 |
predicted gene 14211 |
18111 |
0.18 |
| chr9_62040306_62040468 | 0.13 |
Paqr5 |
progestin and adipoQ receptor family member V |
13531 |
0.2 |
| chr13_34054480_34054653 | 0.13 |
Bphl |
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
5361 |
0.12 |
| chr8_121108318_121108477 | 0.13 |
Mthfsd |
methenyltetrahydrofolate synthetase domain containing |
5 |
0.96 |
| chr5_90366520_90366679 | 0.13 |
Gm9958 |
predicted gene 9958 |
21 |
0.5 |
| chr9_48819032_48819183 | 0.13 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
16838 |
0.22 |
| chr14_25458404_25458994 | 0.13 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
486 |
0.55 |
| chr2_167834679_167834867 | 0.13 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
1127 |
0.46 |
| chr9_124424253_124424419 | 0.13 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
182 |
0.93 |
| chr9_111156088_111156255 | 0.13 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
5132 |
0.2 |
| chr2_30124472_30124630 | 0.12 |
Zer1 |
zyg-11 related, cell cycle regulator |
28 |
0.95 |
| chr17_10040936_10041349 | 0.12 |
Gm49809 |
predicted gene, 49809 |
93305 |
0.08 |
| chr18_10532849_10533008 | 0.12 |
Gm24894 |
predicted gene, 24894 |
18478 |
0.16 |
| chr18_39491582_39491748 | 0.12 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
364 |
0.91 |
| chr17_86662584_86662735 | 0.12 |
Gm18832 |
predicted gene, 18832 |
22837 |
0.19 |
| chr12_110446883_110447049 | 0.12 |
Ppp2r5c |
protein phosphatase 2, regulatory subunit B', gamma |
154 |
0.95 |
| chr3_83286962_83287132 | 0.12 |
Gm38096 |
predicted gene, 38096 |
116355 |
0.06 |
| chr18_21300026_21300190 | 0.12 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
15 |
0.97 |
| chr17_26561346_26561508 | 0.12 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
62 |
0.5 |
| chr2_125859018_125859178 | 0.12 |
Galk2 |
galactokinase 2 |
11 |
0.51 |
| chr17_45619482_45619671 | 0.12 |
Gm25008 |
predicted gene, 25008 |
1103 |
0.32 |
| chr1_72641063_72641244 | 0.12 |
Gm39662 |
predicted gene, 39662 |
35 |
0.97 |
| chr5_150611809_150611960 | 0.12 |
Gm43597 |
predicted gene 43597 |
4881 |
0.11 |
| chr6_94813223_94813696 | 0.12 |
Gm7833 |
predicted gene 7833 |
7071 |
0.15 |
| chr10_80433895_80434247 | 0.12 |
Tcf3 |
transcription factor 3 |
424 |
0.68 |
| chr2_90579423_90579643 | 0.12 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
1114 |
0.56 |
| chr14_52019661_52019820 | 0.12 |
Zfp219 |
zinc finger protein 219 |
27 |
0.81 |
| chr12_111574326_111574506 | 0.12 |
2810029C07Rik |
RIKEN cDNA 2810029C07 gene |
14 |
0.59 |
| chr8_108744763_108744935 | 0.12 |
Gm38042 |
predicted gene, 38042 |
7256 |
0.25 |
| chr11_115433171_115433606 | 0.12 |
Kctd2 |
potassium channel tetramerisation domain containing 2 |
4035 |
0.09 |
| chr13_32338696_32338889 | 0.12 |
A730091E23Rik |
RIKEN cDNA A730091E23 gene |
27 |
0.52 |
| chr11_101646100_101646269 | 0.12 |
Gm23971 |
predicted gene, 23971 |
254 |
0.82 |
| chr14_55660395_55660554 | 0.11 |
Mdp1 |
magnesium-dependent phosphatase 1 |
23 |
0.9 |
| chr19_24225064_24225259 | 0.11 |
Tjp2 |
tight junction protein 2 |
131 |
0.96 |
| chr13_113509396_113509559 | 0.11 |
4921509O07Rik |
RIKEN cDNA 4921509O07 gene |
6633 |
0.17 |
| chr5_86172820_86173003 | 0.11 |
Uba6 |
ubiquitin-like modifier activating enzyme 6 |
108 |
0.97 |
| chrX_101222593_101222760 | 0.11 |
Snx12 |
sorting nexin 12 |
113 |
0.94 |
| chr5_38483324_38483475 | 0.11 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
14 |
0.98 |
| chr2_181599163_181599377 | 0.11 |
Samd10 |
sterile alpha motif domain containing 10 |
58 |
0.93 |
| chr14_61556616_61556767 | 0.11 |
Spryd7 |
SPRY domain containing 7 |
67 |
0.94 |
| chr10_68274977_68275295 | 0.11 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
3585 |
0.28 |
| chr4_138304765_138304921 | 0.11 |
Ddost |
dolichyl-di-phosphooligosaccharide-protein glycotransferase |
113 |
0.94 |
| chr7_140999352_140999505 | 0.11 |
Gm7514 |
predicted gene 7514 |
5915 |
0.08 |
| chr1_90603338_90603497 | 0.11 |
Cops8 |
COP9 signalosome subunit 8 |
59 |
0.98 |
| chr5_114461138_114461289 | 0.11 |
Mvk |
mevalonate kinase |
8793 |
0.15 |
| chr3_51483949_51484322 | 0.11 |
Rab33b |
RAB33B, member RAS oncogene family |
169 |
0.83 |
| chr14_14012284_14012666 | 0.11 |
Atxn7 |
ataxin 7 |
16 |
0.98 |
| chr8_84976570_84976736 | 0.11 |
AC163703.1 |
|
111 |
0.88 |
| chr16_85803386_85803666 | 0.11 |
Adamts1 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
413 |
0.88 |
| chr19_9989208_9989752 | 0.11 |
Best1 |
bestrophin 1 |
3435 |
0.13 |
| chr16_78301611_78301777 | 0.11 |
Cxadr |
coxsackie virus and adenovirus receptor |
62 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 0.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.5 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.2 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.0 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.0 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.2 | GO:0043918 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0043723 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |