Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
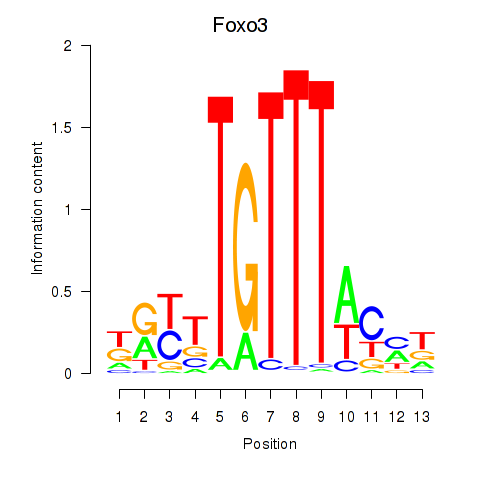
Results for Foxo3
Z-value: 1.23
Transcription factors associated with Foxo3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo3
|
ENSMUSG00000048756.5 | forkhead box O3 |
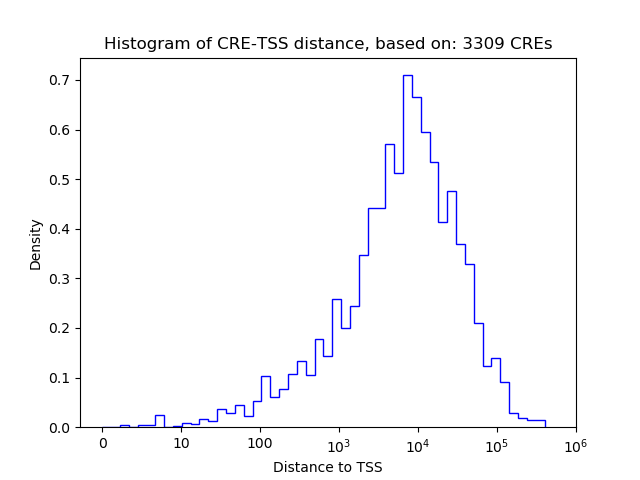
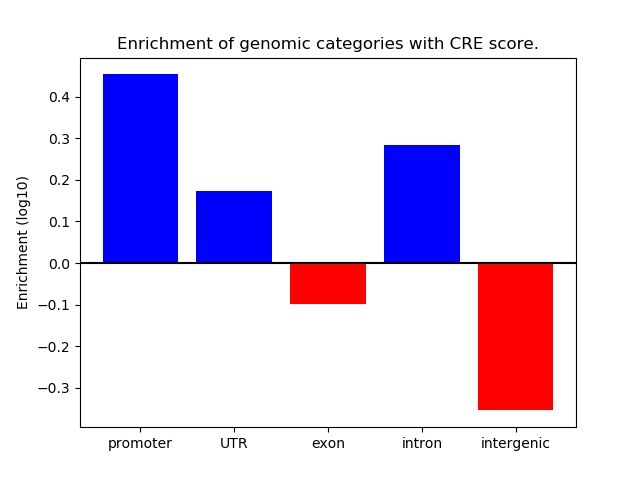
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_42271116_42271452 | Foxo3 | 5412 | 0.273623 | -0.95 | 3.1e-03 | Click! |
| chr10_42226224_42226388 | Foxo3 | 32060 | 0.203915 | 0.95 | 4.0e-03 | Click! |
| chr10_42251679_42251844 | Foxo3 | 6605 | 0.273648 | -0.94 | 6.1e-03 | Click! |
| chr10_42211682_42211846 | Foxo3 | 46602 | 0.162786 | -0.93 | 6.5e-03 | Click! |
| chr10_42233360_42233752 | Foxo3 | 24810 | 0.224152 | -0.93 | 7.4e-03 | Click! |
Activity of the Foxo3 motif across conditions
Conditions sorted by the z-value of the Foxo3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
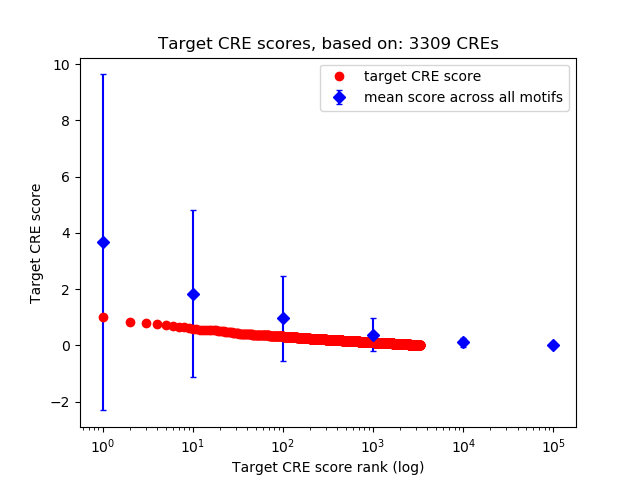
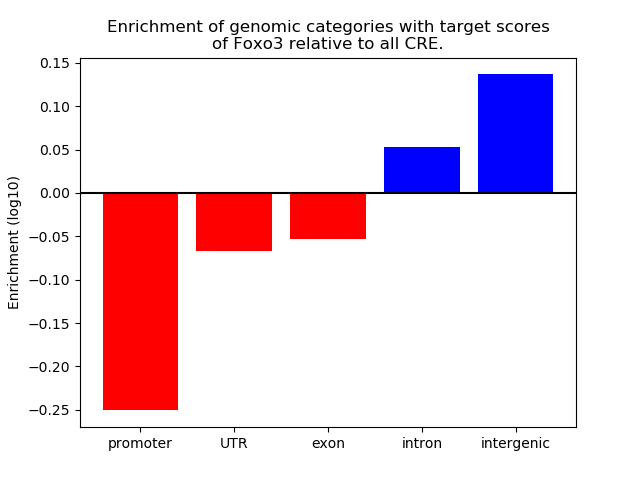
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_25648898_25649067 | 1.00 |
Gm22645 |
predicted gene, 22645 |
45238 |
0.05 |
| chr5_27411249_27411405 | 0.84 |
Speer4b |
spermatogenesis associated glutamate (E)-rich protein 4B |
90035 |
0.09 |
| chr1_184330776_184331172 | 0.78 |
Gm37223 |
predicted gene, 37223 |
27355 |
0.2 |
| chr13_37468607_37468985 | 0.76 |
Gm47731 |
predicted gene, 47731 |
3242 |
0.13 |
| chr3_69778568_69778734 | 0.71 |
Nmd3 |
NMD3 ribosome export adaptor |
32219 |
0.16 |
| chr2_158153808_158154000 | 0.70 |
Tgm2 |
transglutaminase 2, C polypeptide |
7468 |
0.16 |
| chr12_71430131_71430651 | 0.66 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
46183 |
0.12 |
| chr19_6041292_6041476 | 0.64 |
Gm8034 |
predicted gene 8034 |
3438 |
0.07 |
| chr2_25363590_25363748 | 0.62 |
Uap1l1 |
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
29 |
0.93 |
| chr5_8962552_8962757 | 0.60 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
4102 |
0.15 |
| chr18_20920867_20921026 | 0.57 |
Rnf125 |
ring finger protein 125 |
23679 |
0.17 |
| chr17_30010480_30010631 | 0.56 |
Zfand3 |
zinc finger, AN1-type domain 3 |
2758 |
0.19 |
| chr6_57513458_57513615 | 0.55 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
2071 |
0.25 |
| chr2_164161443_164161616 | 0.54 |
n-R5s207 |
nuclear encoded rRNA 5S 207 |
4008 |
0.11 |
| chr10_121125476_121125627 | 0.54 |
Gm48410 |
predicted gene, 48410 |
10306 |
0.16 |
| chr6_17255791_17255960 | 0.54 |
Cav2 |
caveolin 2 |
25310 |
0.17 |
| chr7_35116533_35116684 | 0.53 |
Cebpa |
CCAAT/enhancer binding protein (C/EBP), alpha |
2685 |
0.13 |
| chr13_100532997_100533171 | 0.53 |
Ocln |
occludin |
19376 |
0.11 |
| chr1_58475799_58475950 | 0.52 |
Orc2 |
origin recognition complex, subunit 2 |
5575 |
0.13 |
| chr1_167320339_167320493 | 0.52 |
Tmco1 |
transmembrane and coiled-coil domains 1 |
6279 |
0.12 |
| chr10_116221046_116221335 | 0.50 |
Ptprr |
protein tyrosine phosphatase, receptor type, R |
24794 |
0.18 |
| chr15_55819504_55819678 | 0.49 |
Sntb1 |
syntrophin, basic 1 |
86719 |
0.08 |
| chr13_28654991_28655297 | 0.49 |
Mir6368 |
microRNA 6368 |
55729 |
0.15 |
| chr8_105746150_105746318 | 0.47 |
Gfod2 |
glucose-fructose oxidoreductase domain containing 2 |
12362 |
0.09 |
| chr8_70593155_70593355 | 0.47 |
Isyna1 |
myo-inositol 1-phosphate synthase A1 |
1118 |
0.29 |
| chr13_34301223_34301392 | 0.47 |
Gm47086 |
predicted gene, 47086 |
17501 |
0.18 |
| chr5_121838276_121838548 | 0.46 |
Sh2b3 |
SH2B adaptor protein 3 |
766 |
0.47 |
| chr19_29101814_29101986 | 0.45 |
Rcl1 |
RNA terminal phosphate cyclase-like 1 |
525 |
0.69 |
| chr6_128519043_128519655 | 0.45 |
Pzp |
PZP, alpha-2-macroglobulin like |
7354 |
0.09 |
| chr11_119834463_119834628 | 0.45 |
Rptoros |
regulatory associated protein of MTOR, complex 1, opposite strand |
180 |
0.94 |
| chr6_116747946_116748114 | 0.43 |
Gm22882 |
predicted gene, 22882 |
17622 |
0.16 |
| chr4_43181848_43182012 | 0.42 |
Unc13b |
unc-13 homolog B |
1362 |
0.39 |
| chr8_125337571_125337768 | 0.41 |
Gm16237 |
predicted gene 16237 |
110346 |
0.06 |
| chr14_8124267_8124431 | 0.41 |
Pxk |
PX domain containing serine/threonine kinase |
11005 |
0.18 |
| chr13_54101192_54101366 | 0.41 |
Sfxn1 |
sideroflexin 1 |
8568 |
0.19 |
| chr5_99362991_99363432 | 0.41 |
Gm35394 |
predicted gene, 35394 |
89116 |
0.08 |
| chr8_125381813_125381979 | 0.40 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
110814 |
0.06 |
| chr15_11054061_11054248 | 0.40 |
Adamts12 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 |
10636 |
0.18 |
| chr19_42581997_42582175 | 0.40 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
925 |
0.56 |
| chr5_62858525_62858676 | 0.40 |
Dthd1 |
death domain containing 1 |
44777 |
0.19 |
| chr6_113991518_113991701 | 0.40 |
Gm15083 |
predicted gene 15083 |
13180 |
0.17 |
| chr1_184357776_184358209 | 0.39 |
Gm37223 |
predicted gene, 37223 |
337 |
0.91 |
| chr10_75742534_75742685 | 0.39 |
Cabin1 |
calcineurin binding protein 1 |
8838 |
0.11 |
| chr5_145994754_145994905 | 0.39 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
3186 |
0.17 |
| chr4_126113392_126113559 | 0.38 |
Stk40 |
serine/threonine kinase 40 |
4797 |
0.12 |
| chr15_93515418_93515569 | 0.38 |
Prickle1 |
prickle planar cell polarity protein 1 |
4030 |
0.3 |
| chr11_76849502_76849693 | 0.38 |
Cpd |
carboxypeptidase D |
2579 |
0.29 |
| chr3_102454630_102454805 | 0.38 |
Ngf |
nerve growth factor |
15202 |
0.18 |
| chr4_80931106_80931434 | 0.38 |
Lurap1l |
leucine rich adaptor protein 1-like |
20624 |
0.22 |
| chr16_22439730_22439912 | 0.37 |
Etv5 |
ets variant 5 |
102 |
0.97 |
| chr5_119576958_119577127 | 0.37 |
Tbx3os1 |
T-box 3, opposite strand 1 |
2096 |
0.29 |
| chr5_123201599_123201763 | 0.37 |
Gm43409 |
predicted gene 43409 |
10093 |
0.1 |
| chr12_8643417_8643595 | 0.37 |
Pum2 |
pumilio RNA-binding family member 2 |
30628 |
0.17 |
| chr10_37145852_37146014 | 0.37 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
4844 |
0.21 |
| chr3_142569357_142569508 | 0.37 |
Gbp3 |
guanylate binding protein 3 |
8198 |
0.13 |
| chr17_64643456_64643864 | 0.37 |
Man2a1 |
mannosidase 2, alpha 1 |
42924 |
0.17 |
| chr8_64790589_64790740 | 0.37 |
Klhl2 |
kelch-like 2, Mayven |
9000 |
0.16 |
| chr13_29049878_29050059 | 0.36 |
A330102I10Rik |
RIKEN cDNA A330102I10 gene |
33260 |
0.19 |
| chr6_128523521_128524414 | 0.36 |
Pzp |
PZP, alpha-2-macroglobulin like |
2736 |
0.13 |
| chr7_49358786_49358938 | 0.36 |
Nav2 |
neuron navigator 2 |
5859 |
0.24 |
| chr14_57375309_57375470 | 0.36 |
Gm29717 |
predicted gene, 29717 |
9861 |
0.14 |
| chr13_95894500_95894710 | 0.36 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
2122 |
0.3 |
| chr10_105311004_105311282 | 0.36 |
Gm48206 |
predicted gene, 48206 |
29106 |
0.18 |
| chr15_10183678_10183833 | 0.36 |
Prlr |
prolactin receptor |
5974 |
0.31 |
| chr4_132650270_132650435 | 0.35 |
Eya3 |
EYA transcriptional coactivator and phosphatase 3 |
6348 |
0.17 |
| chr14_27310963_27311149 | 0.35 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
25010 |
0.19 |
| chr10_63226846_63227134 | 0.35 |
Herc4 |
hect domain and RLD 4 |
16820 |
0.11 |
| chr7_12929925_12930160 | 0.35 |
Gm26325 |
predicted gene, 26325 |
732 |
0.4 |
| chr6_71824307_71824486 | 0.35 |
Mrpl35 |
mitochondrial ribosomal protein L35 |
586 |
0.6 |
| chr17_86303142_86303298 | 0.35 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
16042 |
0.26 |
| chr16_22901028_22901197 | 0.35 |
Ahsg |
alpha-2-HS-glycoprotein |
6248 |
0.12 |
| chr5_63936466_63936784 | 0.34 |
Rell1 |
RELT-like 1 |
5711 |
0.18 |
| chr18_53745041_53745208 | 0.34 |
Cep120 |
centrosomal protein 120 |
516 |
0.86 |
| chr19_61029220_61029421 | 0.34 |
Gm22520 |
predicted gene, 22520 |
15775 |
0.19 |
| chr1_166032218_166032391 | 0.34 |
Gm7626 |
predicted gene 7626 |
11387 |
0.15 |
| chr15_82748520_82748679 | 0.34 |
Gm49433 |
predicted gene, 49433 |
6483 |
0.1 |
| chr1_105908668_105908838 | 0.34 |
Gm37779 |
predicted gene, 37779 |
17741 |
0.16 |
| chr18_41861273_41861424 | 0.34 |
Gm50410 |
predicted gene, 50410 |
13486 |
0.22 |
| chr4_95317985_95318146 | 0.34 |
Gm29064 |
predicted gene 29064 |
84725 |
0.08 |
| chr16_14155801_14155952 | 0.34 |
Marf1 |
meiosis regulator and mRNA stability 1 |
3391 |
0.17 |
| chr13_35023472_35023629 | 0.34 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
3544 |
0.15 |
| chr12_108264946_108265126 | 0.33 |
Ccdc85c |
coiled-coil domain containing 85C |
10097 |
0.18 |
| chr6_31317781_31317947 | 0.33 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
4107 |
0.19 |
| chr6_128481760_128482446 | 0.33 |
Pzp |
PZP, alpha-2-macroglobulin like |
5752 |
0.09 |
| chr3_142757820_142758243 | 0.33 |
Gtf2b |
general transcription factor IIB |
7014 |
0.12 |
| chr6_16701972_16702148 | 0.33 |
Gm36669 |
predicted gene, 36669 |
75464 |
0.11 |
| chr13_100849368_100849536 | 0.33 |
Gm29502 |
predicted gene 29502 |
2019 |
0.24 |
| chr5_122527903_122528070 | 0.33 |
Gm22965 |
predicted gene, 22965 |
21197 |
0.09 |
| chr13_91749969_91750270 | 0.33 |
Gm27656 |
predicted gene, 27656 |
1987 |
0.26 |
| chr10_61160789_61160964 | 0.33 |
Tbata |
thymus, brain and testes associated |
11078 |
0.15 |
| chr5_92773855_92774029 | 0.33 |
Mir1961 |
microRNA 1961 |
14620 |
0.18 |
| chr11_35970865_35971211 | 0.32 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
9489 |
0.23 |
| chr2_128172178_128172337 | 0.32 |
Gm14009 |
predicted gene 14009 |
17730 |
0.2 |
| chr1_174907689_174908002 | 0.32 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
13974 |
0.29 |
| chr5_90560915_90561091 | 0.32 |
Albfm1 |
albumin superfamily member 1 |
104 |
0.94 |
| chr10_94573136_94573386 | 0.32 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
1996 |
0.28 |
| chrX_164035471_164035631 | 0.32 |
Car5b |
carbonic anhydrase 5b, mitochondrial |
7554 |
0.2 |
| chr5_145994529_145994680 | 0.32 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
2961 |
0.18 |
| chr1_9584205_9584668 | 0.32 |
Gm6161 |
predicted gene 6161 |
9888 |
0.13 |
| chr6_115730698_115730849 | 0.32 |
Tmem40 |
transmembrane protein 40 |
4178 |
0.13 |
| chr16_18430516_18430667 | 0.31 |
Txnrd2 |
thioredoxin reductase 2 |
1666 |
0.22 |
| chr17_84183727_84183878 | 0.31 |
Gm36279 |
predicted gene, 36279 |
1954 |
0.27 |
| chr3_108640526_108640690 | 0.31 |
Clcc1 |
chloride channel CLIC-like 1 |
13305 |
0.11 |
| chr17_29423486_29423671 | 0.31 |
Gm36199 |
predicted gene, 36199 |
9257 |
0.12 |
| chr6_28681979_28682177 | 0.31 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
22831 |
0.21 |
| chr3_142530130_142530290 | 0.31 |
Gbp7 |
guanylate binding protein 7 |
132 |
0.95 |
| chr10_82753022_82753185 | 0.31 |
Nfyb |
nuclear transcription factor-Y beta |
553 |
0.67 |
| chr3_29732792_29732959 | 0.31 |
Gm37557 |
predicted gene, 37557 |
38409 |
0.19 |
| chr11_80453320_80453471 | 0.30 |
Psmd11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
2973 |
0.25 |
| chr15_57979415_57979570 | 0.30 |
Fam83a |
family with sequence similarity 83, member A |
5927 |
0.18 |
| chr8_84765802_84765975 | 0.30 |
Nfix |
nuclear factor I/X |
7508 |
0.11 |
| chr10_94466409_94466580 | 0.30 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
48363 |
0.13 |
| chr1_162247588_162247739 | 0.30 |
Mir214 |
microRNA 214 |
24295 |
0.14 |
| chr3_157723294_157723469 | 0.30 |
Gm33466 |
predicted gene, 33466 |
3027 |
0.32 |
| chr10_81172506_81172872 | 0.30 |
Eef2 |
eukaryotic translation elongation factor 2 |
3942 |
0.08 |
| chr12_85810078_85810229 | 0.30 |
Erg28 |
ergosterol biosynthesis 28 |
12097 |
0.16 |
| chr13_41250595_41250752 | 0.30 |
Smim13 |
small integral membrane protein 13 |
829 |
0.5 |
| chr11_49088297_49088883 | 0.30 |
Gm12188 |
predicted gene 12188 |
47 |
0.79 |
| chr10_17010158_17010311 | 0.30 |
Gm20125 |
predicted gene, 20125 |
13915 |
0.28 |
| chr17_70797462_70797613 | 0.30 |
Gm41609 |
predicted gene, 41609 |
8460 |
0.13 |
| chr16_26348570_26348826 | 0.30 |
Cldn1 |
claudin 1 |
12096 |
0.29 |
| chr5_124377210_124377362 | 0.29 |
Sbno1 |
strawberry notch 1 |
7523 |
0.12 |
| chr6_94549672_94549849 | 0.29 |
Slc25a26 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
15620 |
0.2 |
| chr1_103048975_103049149 | 0.29 |
Gm22034 |
predicted gene, 22034 |
95514 |
0.09 |
| chr2_62655069_62655412 | 0.29 |
Ifih1 |
interferon induced with helicase C domain 1 |
8985 |
0.18 |
| chr7_141184840_141184997 | 0.29 |
Gm22019 |
predicted gene, 22019 |
3607 |
0.09 |
| chr1_45922514_45922876 | 0.29 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
614 |
0.65 |
| chr8_87121391_87121558 | 0.29 |
Gm27246 |
predicted gene 27246 |
6829 |
0.19 |
| chr13_17717656_17717837 | 0.29 |
Gm48621 |
predicted gene, 48621 |
7882 |
0.12 |
| chr3_98255193_98255424 | 0.29 |
Gm42821 |
predicted gene 42821 |
6709 |
0.15 |
| chr16_33892287_33892604 | 0.29 |
Itgb5 |
integrin beta 5 |
498 |
0.81 |
| chr6_126033747_126033913 | 0.28 |
Ntf3 |
neurotrophin 3 |
131130 |
0.05 |
| chr13_24958493_24958644 | 0.28 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
2714 |
0.2 |
| chr13_4615952_4616121 | 0.28 |
Rpl29-ps2 |
ribosomal protein L29, pseudogene 2 |
1854 |
0.3 |
| chr7_132931708_132932726 | 0.28 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
1020 |
0.34 |
| chr7_141628129_141628296 | 0.28 |
Mir7063 |
microRNA 7063 |
7511 |
0.13 |
| chr16_22694632_22694783 | 0.28 |
Gm8118 |
predicted gene 8118 |
8513 |
0.18 |
| chr12_16870775_16871097 | 0.28 |
Gm36495 |
predicted gene, 36495 |
20055 |
0.13 |
| chr4_99028512_99028838 | 0.28 |
Angptl3 |
angiopoietin-like 3 |
2279 |
0.28 |
| chr14_49703822_49704015 | 0.28 |
Gm16082 |
predicted gene 16082 |
9704 |
0.15 |
| chr1_9787589_9787755 | 0.28 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
3894 |
0.16 |
| chr4_55408726_55408877 | 0.28 |
Gm12513 |
predicted gene 12513 |
506 |
0.73 |
| chr19_4051523_4051732 | 0.28 |
Gstp2 |
glutathione S-transferase, pi 2 |
5604 |
0.07 |
| chr9_61422608_61422759 | 0.28 |
Tle3 |
transducin-like enhancer of split 3 |
12746 |
0.17 |
| chr6_128482799_128482997 | 0.28 |
Pzp |
PZP, alpha-2-macroglobulin like |
4957 |
0.1 |
| chrX_93161079_93161239 | 0.28 |
Gm24123 |
predicted gene, 24123 |
3743 |
0.22 |
| chr3_145790883_145791049 | 0.28 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
19513 |
0.19 |
| chr6_9077922_9078077 | 0.28 |
Gm35736 |
predicted gene, 35736 |
68767 |
0.13 |
| chr10_60349166_60349337 | 0.27 |
Vsir |
V-set immunoregulatory receptor |
72 |
0.97 |
| chr12_110871384_110871543 | 0.27 |
Cinp |
cyclin-dependent kinase 2 interacting protein |
5881 |
0.1 |
| chr18_23496939_23497187 | 0.27 |
Dtna |
dystrobrevin alpha |
228 |
0.96 |
| chr7_68696358_68696602 | 0.27 |
Gm44692 |
predicted gene 44692 |
29987 |
0.18 |
| chr17_56655322_56655554 | 0.27 |
Catsperd |
cation channel sperm associated auxiliary subunit delta |
3656 |
0.13 |
| chr11_94347313_94347820 | 0.27 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
3620 |
0.18 |
| chr5_102751686_102752074 | 0.27 |
Arhgap24 |
Rho GTPase activating protein 24 |
16891 |
0.28 |
| chr8_23284051_23284251 | 0.27 |
Gm24335 |
predicted gene, 24335 |
6013 |
0.16 |
| chr6_29833568_29833845 | 0.27 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
20054 |
0.17 |
| chr1_171786247_171786398 | 0.27 |
Slamf1 |
signaling lymphocytic activation molecule family member 1 |
19167 |
0.11 |
| chr7_135609772_135609956 | 0.27 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
4036 |
0.22 |
| chr7_35127993_35128152 | 0.27 |
Gm45091 |
predicted gene 45091 |
8467 |
0.1 |
| chr2_25156341_25156504 | 0.27 |
Gm13387 |
predicted gene 13387 |
11026 |
0.07 |
| chr19_53890337_53890577 | 0.27 |
Pdcd4 |
programmed cell death 4 |
1774 |
0.29 |
| chr6_86233931_86234108 | 0.27 |
Tgfa |
transforming growth factor alpha |
22425 |
0.13 |
| chr17_56170170_56170436 | 0.26 |
Gm44397 |
predicted gene, 44397 |
1822 |
0.18 |
| chr12_102711766_102711920 | 0.26 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
6913 |
0.1 |
| chr14_114062877_114063028 | 0.26 |
Gm18369 |
predicted gene, 18369 |
109491 |
0.07 |
| chr10_8092375_8092549 | 0.26 |
Gm48614 |
predicted gene, 48614 |
71170 |
0.1 |
| chr15_97590730_97591033 | 0.26 |
Gm49506 |
predicted gene, 49506 |
32591 |
0.19 |
| chr6_5517592_5517751 | 0.26 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
21362 |
0.26 |
| chr1_63178308_63178488 | 0.26 |
Gm26457 |
predicted gene, 26457 |
279 |
0.62 |
| chr2_26314444_26314595 | 0.26 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
996 |
0.38 |
| chr16_95842914_95843065 | 0.26 |
1600002D24Rik |
RIKEN cDNA 1600002D24 gene |
2781 |
0.26 |
| chr13_34239605_34239854 | 0.26 |
Slc22a23 |
solute carrier family 22, member 23 |
46198 |
0.11 |
| chr3_122247143_122247315 | 0.26 |
Gclm |
glutamate-cysteine ligase, modifier subunit |
1148 |
0.3 |
| chr17_29502998_29503149 | 0.26 |
Pim1 |
proviral integration site 1 |
9666 |
0.1 |
| chr2_166035845_166036012 | 0.26 |
Ncoa3 |
nuclear receptor coactivator 3 |
11990 |
0.16 |
| chr10_128271588_128271831 | 0.26 |
Stat2 |
signal transducer and activator of transcription 2 |
1133 |
0.24 |
| chr8_40659245_40659433 | 0.26 |
Adam24 |
a disintegrin and metallopeptidase domain 24 (testase 1) |
15738 |
0.13 |
| chr18_44623567_44623739 | 0.26 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
38012 |
0.17 |
| chr11_107121702_107121874 | 0.26 |
Bptf |
bromodomain PHD finger transcription factor |
10134 |
0.16 |
| chr2_155059266_155059435 | 0.26 |
a |
nonagouti |
11735 |
0.13 |
| chr7_87235141_87235326 | 0.26 |
Gm6230 |
predicted gene 6230 |
7652 |
0.23 |
| chr5_135118666_135118877 | 0.26 |
Gm43500 |
predicted gene 43500 |
4522 |
0.11 |
| chr11_78418219_78418374 | 0.25 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
3921 |
0.12 |
| chr16_91466662_91466825 | 0.25 |
Gm49626 |
predicted gene, 49626 |
1618 |
0.17 |
| chr9_118928821_118928982 | 0.25 |
Ctdspl |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
966 |
0.46 |
| chr13_91759904_91760091 | 0.25 |
Gm27656 |
predicted gene, 27656 |
7891 |
0.15 |
| chr12_18606247_18606408 | 0.25 |
Gm48398 |
predicted gene, 48398 |
4057 |
0.19 |
| chr15_58747025_58747176 | 0.25 |
Gm20712 |
predicted gene 20712 |
34851 |
0.15 |
| chr4_54905681_54905846 | 0.25 |
Zfp462 |
zinc finger protein 462 |
39285 |
0.17 |
| chr1_36075710_36075890 | 0.25 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
7400 |
0.15 |
| chr2_170175810_170175971 | 0.25 |
Zfp217 |
zinc finger protein 217 |
27787 |
0.23 |
| chr15_6709205_6709602 | 0.25 |
Rictor |
RPTOR independent companion of MTOR, complex 2 |
1020 |
0.58 |
| chr12_86476167_86476343 | 0.25 |
Esrrb |
estrogen related receptor, beta |
6138 |
0.27 |
| chr15_31062621_31062832 | 0.25 |
4930430F21Rik |
RIKEN cDNA 4930430F21 gene |
23598 |
0.19 |
| chr1_37491813_37491971 | 0.25 |
Gm43213 |
predicted gene 43213 |
2910 |
0.19 |
| chr6_120580202_120580372 | 0.24 |
Gm44124 |
predicted gene, 44124 |
111 |
0.95 |
| chr18_39430680_39430864 | 0.24 |
Gm15337 |
predicted gene 15337 |
41347 |
0.15 |
| chr13_35616741_35616920 | 0.24 |
Gm48707 |
predicted gene, 48707 |
43 |
0.98 |
| chr8_88983561_88983712 | 0.24 |
Mir8110 |
microRNA 8110 |
41099 |
0.17 |
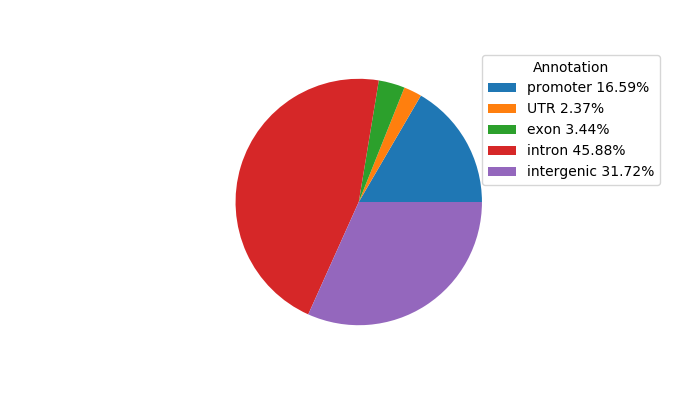
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.2 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.3 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:1903392 | negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.0 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043338 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |