Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
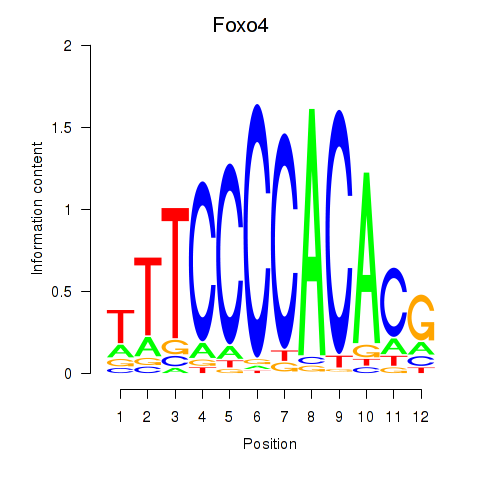
Results for Foxo4
Z-value: 1.66
Transcription factors associated with Foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo4
|
ENSMUSG00000042903.7 | forkhead box O4 |
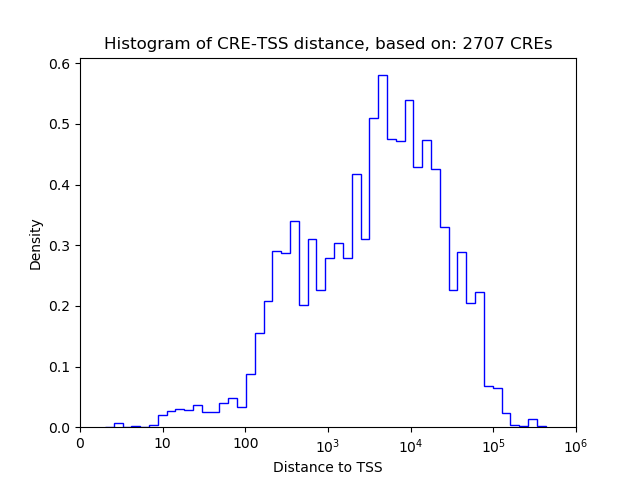
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_101254026_101254193 | Foxo4 | 419 | 0.685478 | -0.88 | 2.0e-02 | Click! |
| chrX_101253687_101253886 | Foxo4 | 742 | 0.448899 | 0.72 | 1.1e-01 | Click! |
| chrX_101254344_101254650 | Foxo4 | 31 | 0.946488 | -0.46 | 3.6e-01 | Click! |
Activity of the Foxo4 motif across conditions
Conditions sorted by the z-value of the Foxo4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
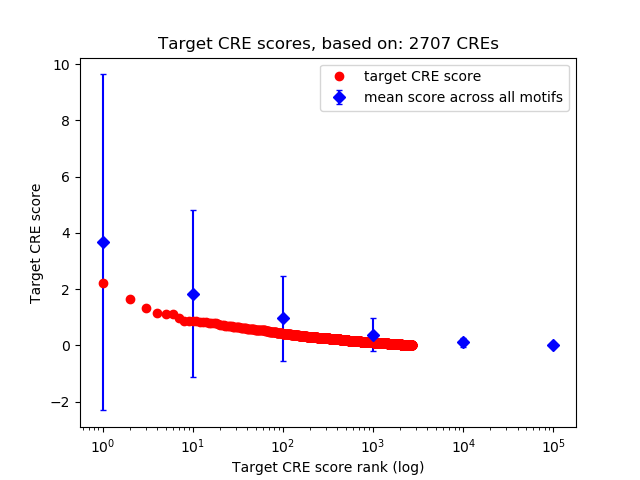
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_80974932_80975087 | 2.23 |
Gm11416 |
predicted gene 11416 |
71785 |
0.1 |
| chr17_46052256_46052407 | 1.63 |
Vegfa |
vascular endothelial growth factor A |
19962 |
0.13 |
| chr16_81353749_81353900 | 1.34 |
Gm49555 |
predicted gene, 49555 |
63662 |
0.13 |
| chr15_62204050_62204201 | 1.16 |
Pvt1 |
Pvt1 oncogene |
14098 |
0.23 |
| chr1_51771064_51771215 | 1.12 |
Myo1b |
myosin IB |
260 |
0.93 |
| chr6_120971997_120972148 | 1.11 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
10646 |
0.14 |
| chr7_98404310_98404465 | 0.97 |
Gm44507 |
predicted gene 44507 |
11237 |
0.14 |
| chr12_32764690_32765098 | 0.87 |
Nampt |
nicotinamide phosphoribosyltransferase |
54651 |
0.11 |
| chr9_106231818_106232229 | 0.87 |
Alas1 |
aminolevulinic acid synthase 1 |
5061 |
0.11 |
| chr8_121970329_121970503 | 0.86 |
Gm17709 |
predicted gene, 17709 |
3406 |
0.13 |
| chr1_21248546_21248733 | 0.86 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4882 |
0.13 |
| chr16_37901578_37901755 | 0.84 |
Gpr156 |
G protein-coupled receptor 156 |
14830 |
0.14 |
| chr7_3630090_3630358 | 0.83 |
Prpf31 |
pre-mRNA processing factor 31 |
165 |
0.62 |
| chr7_87367938_87368104 | 0.82 |
Nox4 |
NADPH oxidase 4 |
121209 |
0.05 |
| chr13_24378651_24378989 | 0.81 |
Cmah |
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
2737 |
0.19 |
| chr8_93185590_93185741 | 0.79 |
Gm45909 |
predicted gene 45909 |
5693 |
0.14 |
| chr16_4414348_4414533 | 0.79 |
Adcy9 |
adenylate cyclase 9 |
5147 |
0.24 |
| chr12_30200282_30200708 | 0.79 |
Sntg2 |
syntrophin, gamma 2 |
840 |
0.72 |
| chr17_27621069_27621255 | 0.78 |
Nudt3 |
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
1231 |
0.25 |
| chr9_123153278_123153441 | 0.74 |
Clec3b |
C-type lectin domain family 3, member b |
2413 |
0.18 |
| chr7_98351265_98351558 | 0.73 |
Tsku |
tsukushi, small leucine rich proteoglycan |
8668 |
0.17 |
| chr3_133725268_133725422 | 0.72 |
Gm40153 |
predicted gene, 40153 |
61954 |
0.1 |
| chr9_122164867_122165083 | 0.70 |
Snrk |
SNF related kinase |
1106 |
0.4 |
| chr2_177468355_177468573 | 0.70 |
Zfp970 |
zinc finger protein 970 |
3618 |
0.19 |
| chr17_10310410_10310561 | 0.70 |
Qk |
quaking |
8876 |
0.24 |
| chr8_70832381_70832563 | 0.69 |
Arrdc2 |
arrestin domain containing 2 |
4115 |
0.09 |
| chr8_66494561_66494830 | 0.68 |
Gm32568 |
predicted gene, 32568 |
364 |
0.85 |
| chr16_77405236_77405411 | 0.67 |
Gm21816 |
predicted gene, 21816 |
4476 |
0.13 |
| chr3_107793106_107793505 | 0.67 |
Gm43233 |
predicted gene 43233 |
11449 |
0.12 |
| chr7_119855181_119855332 | 0.66 |
Dcun1d3 |
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
6548 |
0.15 |
| chr12_76369660_76370048 | 0.65 |
Zbtb25 |
zinc finger and BTB domain containing 25 |
252 |
0.62 |
| chr8_93195399_93195725 | 0.65 |
Ces1d |
carboxylesterase 1D |
2215 |
0.21 |
| chr6_108426324_108426499 | 0.63 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
8534 |
0.22 |
| chr3_96630277_96630623 | 0.62 |
Rbm8a |
RNA binding motif protein 8a |
253 |
0.67 |
| chr6_115668757_115668908 | 0.61 |
Raf1 |
v-raf-leukemia viral oncogene 1 |
7132 |
0.12 |
| chr7_124491258_124491413 | 0.61 |
Gm23168 |
predicted gene, 23168 |
1571 |
0.43 |
| chr9_103301161_103301484 | 0.60 |
1300017J02Rik |
RIKEN cDNA 1300017J02 gene |
3760 |
0.19 |
| chr10_77629279_77629435 | 0.60 |
Ube2g2 |
ubiquitin-conjugating enzyme E2G 2 |
6979 |
0.08 |
| chr6_127044968_127045119 | 0.60 |
Fgf23 |
fibroblast growth factor 23 |
27859 |
0.1 |
| chr17_84137478_84137775 | 0.60 |
Gm19696 |
predicted gene, 19696 |
112 |
0.96 |
| chr6_42360337_42360550 | 0.59 |
Zyx |
zyxin |
5306 |
0.09 |
| chr15_3487391_3487651 | 0.58 |
Ghr |
growth hormone receptor |
15877 |
0.27 |
| chr5_146246268_146246419 | 0.58 |
Gm15739 |
predicted gene 15739 |
2208 |
0.21 |
| chr1_59159262_59159448 | 0.58 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
687 |
0.57 |
| chr8_46918583_46918754 | 0.58 |
1700011L03Rik |
RIKEN cDNA 1700011L03 gene |
30131 |
0.16 |
| chr1_21263057_21263217 | 0.57 |
Gm28836 |
predicted gene 28836 |
8456 |
0.11 |
| chr17_46454562_46454932 | 0.57 |
Gm5093 |
predicted gene 5093 |
14650 |
0.09 |
| chr15_58942719_58942985 | 0.56 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
9044 |
0.13 |
| chr12_71941808_71941989 | 0.56 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
52168 |
0.13 |
| chr12_8785591_8785796 | 0.56 |
Sdc1 |
syndecan 1 |
13890 |
0.17 |
| chr17_75539486_75539637 | 0.56 |
Fam98a |
family with sequence similarity 98, member A |
1011 |
0.65 |
| chr7_126667358_126667803 | 0.56 |
Sgf29 |
SAGA complex associated factor 29 |
3787 |
0.08 |
| chr10_121349997_121350168 | 0.55 |
Gm48435 |
predicted gene, 48435 |
378 |
0.73 |
| chr6_31260546_31260737 | 0.55 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
17207 |
0.15 |
| chr16_43236306_43236477 | 0.55 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
352 |
0.9 |
| chr6_148945956_148946305 | 0.55 |
Sinhcaf |
SIN3-HDAC complex associated factor |
25 |
0.94 |
| chr10_23851933_23852432 | 0.55 |
Vnn3 |
vanin 3 |
720 |
0.56 |
| chr16_77351638_77351789 | 0.54 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
22209 |
0.13 |
| chr4_122955991_122956232 | 0.54 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
3879 |
0.16 |
| chr9_103213949_103214100 | 0.53 |
Trf |
transferrin |
2018 |
0.29 |
| chr11_69367124_69367290 | 0.53 |
Chd3 |
chromodomain helicase DNA binding protein 3 |
2184 |
0.13 |
| chr19_53898302_53898495 | 0.53 |
Pdcd4 |
programmed cell death 4 |
4568 |
0.17 |
| chr10_3114251_3114403 | 0.53 |
Gm3318 |
predicted gene 3318 |
4660 |
0.15 |
| chr1_88137698_88137920 | 0.53 |
Ugt1a6a |
UDP glucuronosyltransferase 1 family, polypeptide A6A |
569 |
0.47 |
| chr13_67194609_67194806 | 0.53 |
Zfp455 |
zinc finger protein 455 |
172 |
0.88 |
| chr7_134381581_134381749 | 0.52 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
3996 |
0.35 |
| chr11_120807385_120807536 | 0.52 |
Fasn |
fatty acid synthase |
1145 |
0.28 |
| chr9_25592470_25592621 | 0.51 |
Gm25346 |
predicted gene, 25346 |
22740 |
0.2 |
| chr6_94669947_94670290 | 0.51 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
8747 |
0.23 |
| chr13_102593887_102594038 | 0.51 |
Gm29927 |
predicted gene, 29927 |
3677 |
0.27 |
| chr1_193934263_193934432 | 0.49 |
Gm21362 |
predicted gene, 21362 |
67338 |
0.14 |
| chr5_130009254_130009405 | 0.49 |
Asl |
argininosuccinate lyase |
5393 |
0.12 |
| chr5_28045123_28045274 | 0.49 |
Gm43611 |
predicted gene 43611 |
8792 |
0.16 |
| chr18_32432396_32432575 | 0.48 |
Bin1 |
bridging integrator 1 |
4451 |
0.19 |
| chr3_118601540_118602026 | 0.48 |
Dpyd |
dihydropyrimidine dehydrogenase |
39597 |
0.15 |
| chr17_46468128_46468340 | 0.48 |
Gm47119 |
predicted gene, 47119 |
8273 |
0.1 |
| chr2_25292972_25293141 | 0.48 |
Grin1os |
glutamate receptor, ionotropic, NMDA1 (zeta 1), opposite strand |
1836 |
0.11 |
| chr7_79273801_79273960 | 0.48 |
Abhd2 |
abhydrolase domain containing 2 |
626 |
0.48 |
| chr1_88102885_88103050 | 0.47 |
Ugt1a7c |
UDP glucuronosyltransferase 1 family, polypeptide A7C |
285 |
0.43 |
| chr2_122244595_122244916 | 0.47 |
Sord |
sorbitol dehydrogenase |
10006 |
0.11 |
| chr4_6265892_6266043 | 0.47 |
Gm11798 |
predicted gene 11798 |
5002 |
0.21 |
| chr9_35267693_35268345 | 0.47 |
Rpusd4 |
RNA pseudouridylate synthase domain containing 4 |
154 |
0.57 |
| chr7_44207877_44208028 | 0.47 |
Klk1b4 |
kallikrein 1-related pepidase b4 |
517 |
0.41 |
| chr8_70234885_70235099 | 0.46 |
Sugp2 |
SURP and G patch domain containing 2 |
369 |
0.55 |
| chr10_80958504_80958805 | 0.45 |
Gm3828 |
predicted gene 3828 |
3944 |
0.12 |
| chr3_129744894_129745074 | 0.45 |
Gm42650 |
predicted gene 42650 |
356 |
0.81 |
| chr7_120843161_120843378 | 0.45 |
Eef2k |
eukaryotic elongation factor-2 kinase |
298 |
0.86 |
| chr19_44044543_44044826 | 0.45 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
15476 |
0.13 |
| chr1_67192197_67192413 | 0.45 |
Gm15668 |
predicted gene 15668 |
56895 |
0.12 |
| chr11_11961796_11961953 | 0.44 |
Grb10 |
growth factor receptor bound protein 10 |
5850 |
0.23 |
| chr18_12224214_12224414 | 0.44 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
227 |
0.91 |
| chr6_54251791_54251942 | 0.44 |
Gm15527 |
predicted gene 15527 |
3045 |
0.25 |
| chr19_32956944_32957156 | 0.44 |
Gm36860 |
predicted gene, 36860 |
23013 |
0.24 |
| chr7_99173210_99173446 | 0.44 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
3222 |
0.17 |
| chr6_129476147_129476311 | 0.44 |
Clec7a |
C-type lectin domain family 7, member a |
3450 |
0.12 |
| chr2_172449892_172450043 | 0.43 |
Rtf2 |
replication termination factor 2 |
3010 |
0.17 |
| chr5_24976790_24976954 | 0.43 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
8970 |
0.18 |
| chr15_83564154_83564331 | 0.43 |
Tspo |
translocator protein |
367 |
0.52 |
| chr1_87764160_87764454 | 0.42 |
Atg16l1 |
autophagy related 16-like 1 (S. cerevisiae) |
2594 |
0.19 |
| chr10_69258899_69259108 | 0.42 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
6634 |
0.21 |
| chr9_59617318_59618012 | 0.42 |
Parp6 |
poly (ADP-ribose) polymerase family, member 6 |
285 |
0.88 |
| chr19_5796124_5796526 | 0.42 |
Malat1 |
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
627 |
0.48 |
| chr7_90080690_90080897 | 0.42 |
Gm5341 |
predicted pseudogene 5341 |
27175 |
0.1 |
| chr4_150393280_150393601 | 0.42 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
12954 |
0.22 |
| chr10_69214299_69214646 | 0.41 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
830 |
0.51 |
| chr1_62053011_62053191 | 0.41 |
Gm29641 |
predicted gene 29641 |
42238 |
0.17 |
| chr14_14789749_14789914 | 0.41 |
Slc4a7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
3904 |
0.26 |
| chr9_74892937_74893252 | 0.41 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr9_107303087_107303371 | 0.41 |
Gm17041 |
predicted gene 17041 |
1391 |
0.24 |
| chr6_47758577_47758728 | 0.41 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
27722 |
0.12 |
| chr5_40691768_40691967 | 0.41 |
Gm23022 |
predicted gene, 23022 |
285938 |
0.01 |
| chr8_73953761_73954148 | 0.40 |
Gm7948 |
predicted gene 7948 |
82261 |
0.11 |
| chr7_19573353_19573652 | 0.40 |
Gemin7 |
gem nuclear organelle associated protein 7 |
117 |
0.92 |
| chr7_99130459_99130779 | 0.40 |
Gm44975 |
predicted gene 44975 |
7160 |
0.13 |
| chr2_27694949_27695100 | 0.40 |
Rxra |
retinoid X receptor alpha |
14268 |
0.25 |
| chr11_115815536_115815882 | 0.40 |
Tsen54 |
tRNA splicing endonuclease subunit 54 |
11 |
0.95 |
| chr2_38225178_38225332 | 0.40 |
Gm44455 |
predicted gene, 44455 |
133 |
0.96 |
| chr10_68093985_68094164 | 0.40 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
42552 |
0.15 |
| chr1_133712710_133712943 | 0.39 |
Atp2b4 |
ATPase, Ca++ transporting, plasma membrane 4 |
5103 |
0.15 |
| chr11_112038554_112038713 | 0.39 |
Gm11679 |
predicted gene 11679 |
4945 |
0.32 |
| chr7_140104126_140104290 | 0.39 |
Fuom |
fucose mutarotase |
1767 |
0.18 |
| chr10_124590350_124590536 | 0.39 |
4930503E24Rik |
RIKEN cDNA 4930503E24 gene |
67005 |
0.14 |
| chr11_120560412_120560649 | 0.38 |
P4hb |
prolyl 4-hydroxylase, beta polypeptide |
706 |
0.37 |
| chr5_125528257_125528432 | 0.38 |
Tmem132b |
transmembrane protein 132B |
3430 |
0.21 |
| chr9_96983617_96983972 | 0.38 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
401 |
0.83 |
| chr11_78437291_78437469 | 0.38 |
Gm11195 |
predicted gene 11195 |
6727 |
0.1 |
| chr2_70642763_70643161 | 0.38 |
Gorasp2 |
golgi reassembly stacking protein 2 |
18614 |
0.14 |
| chr6_47772904_47773093 | 0.37 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
13376 |
0.13 |
| chr5_123226541_123226692 | 0.37 |
Psmd9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
1574 |
0.24 |
| chr10_81414192_81414360 | 0.37 |
Mir1191b |
microRNA 1191b |
2021 |
0.11 |
| chr9_108089036_108089891 | 0.37 |
Apeh |
acylpeptide hydrolase |
943 |
0.28 |
| chr3_118606217_118606485 | 0.37 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr19_20626512_20626720 | 0.37 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
24655 |
0.19 |
| chr12_32488584_32488761 | 0.37 |
Gm17820 |
predicted gene, 17820 |
1254 |
0.55 |
| chr11_69192936_69193104 | 0.36 |
Alox8 |
arachidonate 8-lipoxygenase |
4364 |
0.12 |
| chr13_101914612_101914792 | 0.36 |
Gm17832 |
predicted gene, 17832 |
5718 |
0.26 |
| chr2_26931955_26932305 | 0.36 |
Surf4 |
surfeit gene 4 |
1253 |
0.2 |
| chr2_30268240_30268600 | 0.36 |
Phyhd1 |
phytanoyl-CoA dioxygenase domain containing 1 |
1557 |
0.21 |
| chr6_125309060_125309481 | 0.36 |
Ltbr |
lymphotoxin B receptor |
295 |
0.81 |
| chr2_160646625_160646797 | 0.36 |
Top1 |
topoisomerase (DNA) I |
731 |
0.65 |
| chr6_47664591_47664742 | 0.36 |
Gm44141 |
predicted gene, 44141 |
29369 |
0.16 |
| chr11_113140051_113140245 | 0.36 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
32929 |
0.22 |
| chr14_30887527_30887997 | 0.36 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
1241 |
0.32 |
| chr8_40564488_40564639 | 0.36 |
Vps37a |
vacuolar protein sorting 37A |
21139 |
0.15 |
| chr10_79897700_79897893 | 0.36 |
Med16 |
mediator complex subunit 16 |
1889 |
0.11 |
| chr9_44083899_44084207 | 0.35 |
Usp2 |
ubiquitin specific peptidase 2 |
886 |
0.32 |
| chr4_150869904_150870096 | 0.35 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
14927 |
0.13 |
| chr11_88512888_88513073 | 0.35 |
Msi2 |
musashi RNA-binding protein 2 |
77167 |
0.09 |
| chr18_33337551_33337772 | 0.35 |
Gm5503 |
predicted gene 5503 |
47294 |
0.17 |
| chr2_132263417_132263641 | 0.35 |
Cds2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
288 |
0.88 |
| chr15_55117700_55118060 | 0.35 |
Gm9920 |
predicted gene 9920 |
4203 |
0.17 |
| chr13_56134012_56134235 | 0.34 |
Macroh2a1 |
macroH2A.1 histone |
702 |
0.66 |
| chr16_24902438_24902606 | 0.34 |
Gm22672 |
predicted gene, 22672 |
18349 |
0.23 |
| chr10_77587179_77587612 | 0.34 |
Pttg1ip |
pituitary tumor-transforming 1 interacting protein |
1999 |
0.18 |
| chr18_56414797_56414976 | 0.34 |
Gramd3 |
GRAM domain containing 3 |
4284 |
0.23 |
| chr6_92183686_92183906 | 0.34 |
Mrps25 |
mitochondrial ribosomal protein S25 |
175 |
0.93 |
| chr4_8733319_8733778 | 0.34 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
17890 |
0.25 |
| chr14_67055418_67055773 | 0.34 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
12066 |
0.16 |
| chr5_77331140_77331293 | 0.34 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
5326 |
0.15 |
| chr3_98013087_98013258 | 0.34 |
Notch2 |
notch 2 |
355 |
0.87 |
| chr1_164559930_164560081 | 0.33 |
D630023O14Rik |
RIKEN cDNA D630023O14 gene |
15450 |
0.17 |
| chr1_13639776_13639929 | 0.33 |
Lactb2 |
lactamase, beta 2 |
1441 |
0.37 |
| chr19_41656077_41656246 | 0.33 |
Slit1 |
slit guidance ligand 1 |
47631 |
0.13 |
| chr14_18232288_18232439 | 0.33 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
6648 |
0.15 |
| chr6_142345778_142346191 | 0.33 |
Pyroxd1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
306 |
0.86 |
| chr9_65188991_65189142 | 0.33 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
6287 |
0.13 |
| chr2_91072228_91072550 | 0.33 |
Slc39a13 |
solute carrier family 39 (metal ion transporter), member 13 |
1972 |
0.19 |
| chr19_40349126_40349331 | 0.33 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
8634 |
0.21 |
| chr6_47651586_47651824 | 0.33 |
Gm44141 |
predicted gene, 44141 |
16408 |
0.19 |
| chr4_109122898_109123075 | 0.33 |
Osbpl9 |
oxysterol binding protein-like 9 |
4950 |
0.24 |
| chr19_47451572_47451805 | 0.32 |
Sh3pxd2a |
SH3 and PX domains 2A |
12559 |
0.19 |
| chr18_53291057_53291223 | 0.32 |
Snx24 |
sorting nexing 24 |
45371 |
0.16 |
| chr10_95318232_95318691 | 0.32 |
Gm48882 |
predicted gene, 48882 |
214 |
0.9 |
| chr19_34524558_34524859 | 0.32 |
Lipa |
lysosomal acid lipase A |
2703 |
0.19 |
| chr10_4621590_4621757 | 0.32 |
Esr1 |
estrogen receptor 1 (alpha) |
9652 |
0.24 |
| chr2_128971807_128971990 | 0.32 |
Gm10762 |
predicted gene 10762 |
3854 |
0.12 |
| chr6_47768512_47768677 | 0.32 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
17780 |
0.13 |
| chr6_47740769_47740920 | 0.32 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
45530 |
0.1 |
| chr2_152733626_152733777 | 0.32 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
2550 |
0.16 |
| chr3_144113744_144114175 | 0.31 |
Gm34078 |
predicted gene, 34078 |
21795 |
0.21 |
| chr3_133749142_133749410 | 0.31 |
Gm6135 |
prediticted gene 6135 |
42228 |
0.15 |
| chr12_39373542_39373711 | 0.31 |
Gm47855 |
predicted gene, 47855 |
24930 |
0.22 |
| chr7_128528420_128528640 | 0.31 |
Bag3 |
BCL2-associated athanogene 3 |
4914 |
0.13 |
| chr10_95385978_95386158 | 0.31 |
Socs2 |
suppressor of cytokine signaling 2 |
6899 |
0.14 |
| chr15_87326683_87326854 | 0.31 |
A930027H12Rik |
RIKEN cDNA A930027H12 gene |
83218 |
0.1 |
| chr15_102028397_102029113 | 0.31 |
Krt18 |
keratin 18 |
575 |
0.64 |
| chr10_120227241_120227692 | 0.31 |
Llph |
LLP homolog, long-term synaptic facilitation (Aplysia) |
280 |
0.88 |
| chr5_89422914_89423162 | 0.31 |
Gc |
vitamin D binding protein |
12590 |
0.26 |
| chr7_99163511_99163819 | 0.31 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
6441 |
0.13 |
| chr11_12464170_12464384 | 0.31 |
Cobl |
cordon-bleu WH2 repeat |
475 |
0.89 |
| chr3_95871977_95872183 | 0.30 |
C920021L13Rik |
RIKEN cDNA C920021L13 gene |
549 |
0.36 |
| chr14_103070733_103070991 | 0.30 |
Cln5 |
ceroid-lipofuscinosis, neuronal 5 |
646 |
0.63 |
| chr9_102990799_102990950 | 0.30 |
Slco2a1 |
solute carrier organic anion transporter family, member 2a1 |
2162 |
0.27 |
| chr3_30506438_30507337 | 0.30 |
Mecom |
MDS1 and EVI1 complex locus |
2600 |
0.2 |
| chr12_85824517_85825257 | 0.30 |
Ttll5 |
tubulin tyrosine ligase-like family, member 5 |
81 |
0.8 |
| chr17_28428613_28428954 | 0.30 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr4_103933550_103933744 | 0.30 |
Gm12719 |
predicted gene 12719 |
5894 |
0.25 |
| chr9_71027981_71028199 | 0.30 |
Lipc |
lipase, hepatic |
75864 |
0.09 |
| chr11_115165779_115165997 | 0.30 |
Slc9a3r1 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
2547 |
0.17 |
| chr12_35674441_35674592 | 0.30 |
9130015A21Rik |
RIKEN cDNA 9130015A21 gene |
13 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.5 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.2 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 0.1 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:1902116 | negative regulation of organelle assembly(GO:1902116) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0046464 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.0 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.3 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 1.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.0 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0044038 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.0 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:2001182 | regulation of interleukin-12 secretion(GO:2001182) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0051767 | nitric-oxide synthase biosynthetic process(GO:0051767) regulation of nitric-oxide synthase biosynthetic process(GO:0051769) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0018565 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0018631 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0034892 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | Genes involved in Insulin receptor signalling cascade |
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |