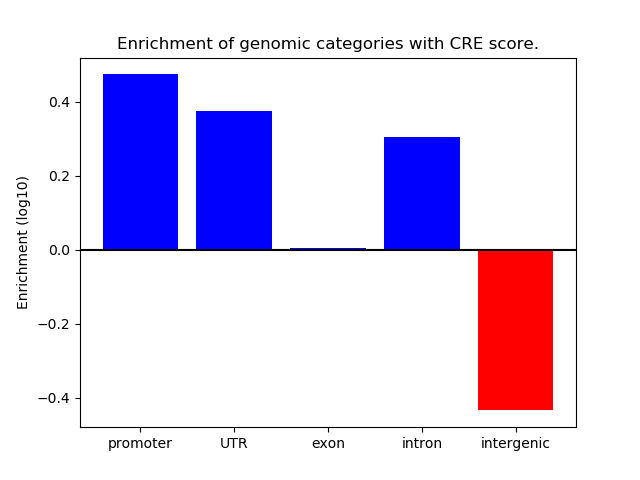
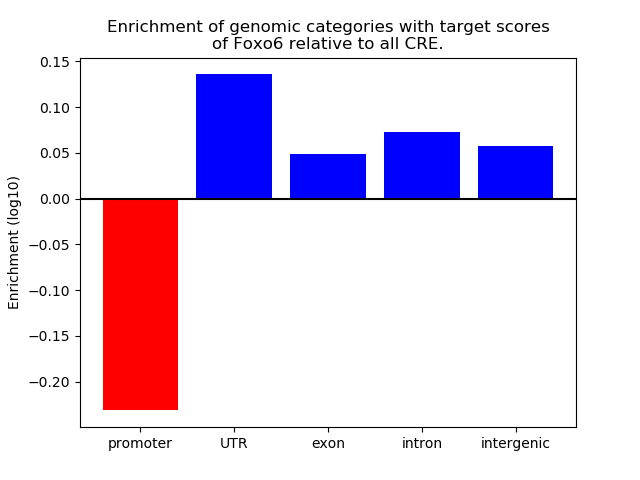
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
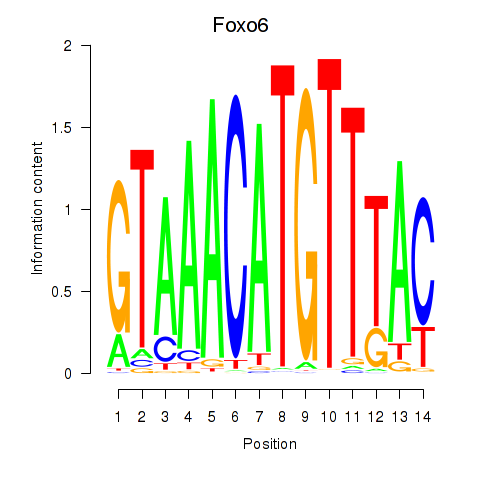
Results for Foxo6
Z-value: 1.26
Transcription factors associated with Foxo6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo6
|
ENSMUSG00000052135.8 | forkhead box O6 |
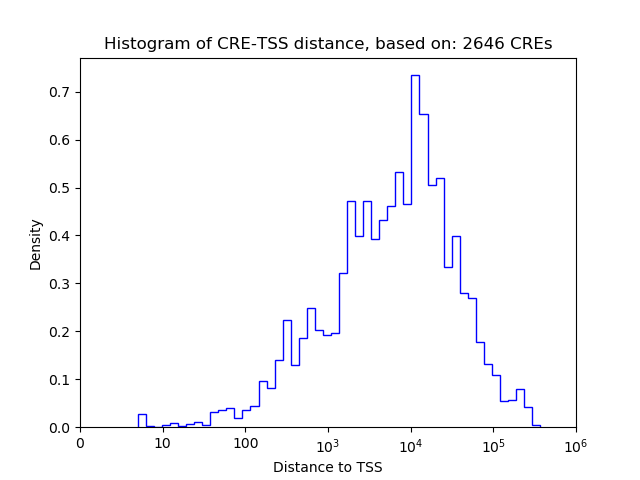
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_120283519_120283670 | Foxo6 | 3755 | 0.255093 | 0.85 | 3.3e-02 | Click! |
| chr4_120248926_120249096 | Foxo6 | 38338 | 0.155782 | -0.82 | 4.4e-02 | Click! |
| chr4_120249226_120249489 | Foxo6 | 37992 | 0.156568 | -0.69 | 1.3e-01 | Click! |
| chr4_120282848_120282999 | Foxo6 | 4426 | 0.242435 | 0.48 | 3.4e-01 | Click! |
| chr4_120283294_120283461 | Foxo6 | 3972 | 0.250350 | -0.36 | 4.9e-01 | Click! |
Activity of the Foxo6 motif across conditions
Conditions sorted by the z-value of the Foxo6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
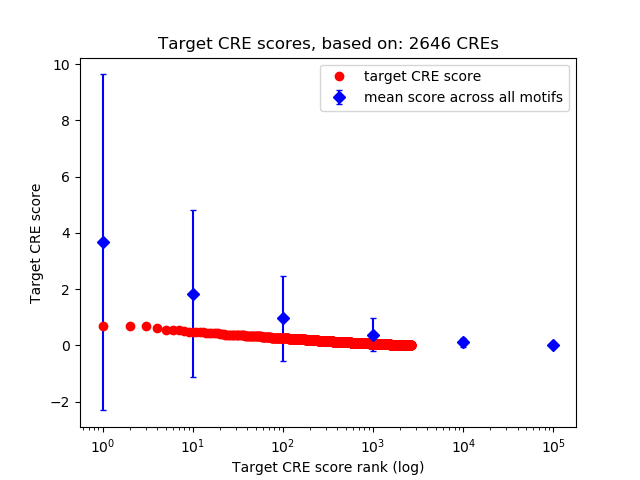
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_180200474_180200787 | 0.68 |
Coq8a |
coenzyme Q8A |
1028 |
0.45 |
| chr8_17783842_17784014 | 0.68 |
Csmd1 |
CUB and Sushi multiple domains 1 |
248342 |
0.02 |
| chr2_143993868_143994250 | 0.67 |
Rrbp1 |
ribosome binding protein 1 |
10894 |
0.18 |
| chr2_143994492_143994838 | 0.63 |
Rrbp1 |
ribosome binding protein 1 |
11500 |
0.18 |
| chr15_87045730_87045890 | 0.56 |
Gm23416 |
predicted gene, 23416 |
23183 |
0.18 |
| chr5_89342174_89342864 | 0.56 |
Gc |
vitamin D binding protein |
93109 |
0.09 |
| chr13_31257761_31257912 | 0.53 |
4930401O12Rik |
RIKEN cDNA 4930401O12 gene |
16750 |
0.19 |
| chr6_95669064_95669249 | 0.50 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
11688 |
0.31 |
| chr4_70410998_70411149 | 0.48 |
Cdk5rap2 |
CDK5 regulatory subunit associated protein 2 |
630 |
0.84 |
| chr6_117598383_117598541 | 0.47 |
Gm45083 |
predicted gene 45083 |
14108 |
0.2 |
| chr15_4836304_4836788 | 0.47 |
Gm49074 |
predicted gene, 49074 |
10841 |
0.18 |
| chr12_4220183_4220334 | 0.47 |
Gm48210 |
predicted gene, 48210 |
1844 |
0.19 |
| chr8_35390018_35390398 | 0.47 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
13548 |
0.15 |
| chr4_80948759_80948910 | 0.46 |
Gm27452 |
predicted gene, 27452 |
18610 |
0.23 |
| chr2_155083519_155083714 | 0.46 |
Gm45609 |
predicted gene 45609 |
1435 |
0.33 |
| chr12_4220651_4220920 | 0.45 |
Gm48210 |
predicted gene, 48210 |
1317 |
0.27 |
| chr10_59786684_59786837 | 0.45 |
Gm17059 |
predicted gene 17059 |
13494 |
0.14 |
| chr10_107289611_107289768 | 0.43 |
Lin7a |
lin-7 homolog A (C. elegans) |
17295 |
0.24 |
| chr7_63922052_63922218 | 0.43 |
Klf13 |
Kruppel-like factor 13 |
2735 |
0.19 |
| chr6_121844056_121844207 | 0.42 |
Mug1 |
murinoglobulin 1 |
3027 |
0.29 |
| chr6_140197945_140198178 | 0.41 |
Gm24174 |
predicted gene, 24174 |
31416 |
0.16 |
| chr9_67830526_67830677 | 0.40 |
C2cd4a |
C2 calcium-dependent domain containing 4A |
1729 |
0.35 |
| chr11_69094794_69095306 | 0.38 |
Per1 |
period circadian clock 1 |
167 |
0.86 |
| chr9_112103407_112103804 | 0.38 |
Mir128-2 |
microRNA 128-2 |
15106 |
0.26 |
| chr5_140129593_140129908 | 0.38 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
14284 |
0.15 |
| chr19_39005964_39006115 | 0.37 |
Cyp2c55 |
cytochrome P450, family 2, subfamily c, polypeptide 55 |
980 |
0.38 |
| chr12_52470605_52470777 | 0.37 |
Gm35135 |
predicted gene, 35135 |
9918 |
0.19 |
| chr9_62355640_62355873 | 0.37 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
10957 |
0.2 |
| chr3_36481808_36481975 | 0.37 |
1810062G17Rik |
RIKEN cDNA 1810062G17 gene |
5954 |
0.12 |
| chr18_12023718_12023871 | 0.37 |
Tmem241 |
transmembrane protein 241 |
50837 |
0.13 |
| chr1_67207008_67207510 | 0.37 |
Gm15668 |
predicted gene 15668 |
41941 |
0.15 |
| chr6_113440512_113440730 | 0.37 |
Gm43928 |
predicted gene, 43928 |
273 |
0.75 |
| chr10_111389057_111389255 | 0.37 |
Gm40761 |
predicted gene, 40761 |
52032 |
0.11 |
| chr19_4878250_4878444 | 0.36 |
Zdhhc24 |
zinc finger, DHHC domain containing 24 |
321 |
0.53 |
| chr6_57543051_57543249 | 0.36 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
7682 |
0.16 |
| chr17_46723600_46724031 | 0.36 |
Gnmt |
glycine N-methyltransferase |
2887 |
0.12 |
| chr9_101182262_101182439 | 0.35 |
Gm38344 |
predicted gene, 38344 |
5558 |
0.13 |
| chr2_155378850_155379039 | 0.35 |
Trp53inp2 |
transformation related protein 53 inducible nuclear protein 2 |
2115 |
0.23 |
| chr10_20486313_20486464 | 0.34 |
Gm17229 |
predicted gene 17229 |
31776 |
0.16 |
| chr9_103217995_103218146 | 0.34 |
Trf |
transferrin |
1796 |
0.31 |
| chr15_9093874_9094049 | 0.34 |
Nadk2 |
NAD kinase 2, mitochondrial |
686 |
0.72 |
| chr4_104768550_104768721 | 0.34 |
C8b |
complement component 8, beta polypeptide |
2240 |
0.4 |
| chr2_59130808_59130980 | 0.34 |
Gm13550 |
predicted gene 13550 |
17729 |
0.19 |
| chr3_144285992_144286165 | 0.33 |
Gm43446 |
predicted gene 43446 |
2267 |
0.32 |
| chr9_95567560_95567711 | 0.33 |
Paqr9 |
progestin and adipoQ receptor family member IX |
7978 |
0.14 |
| chr2_109757612_109757763 | 0.33 |
Gm13932 |
predicted gene 13932 |
34847 |
0.16 |
| chr19_37887154_37887305 | 0.33 |
Myof |
myoferlin |
14274 |
0.19 |
| chr2_31519719_31520357 | 0.33 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr12_79441750_79441941 | 0.32 |
Rad51b |
RAD51 paralog B |
114492 |
0.06 |
| chr5_51799710_51799877 | 0.32 |
Gm43606 |
predicted gene 43606 |
58982 |
0.12 |
| chr8_22858612_22858763 | 0.32 |
Kat6a |
K(lysine) acetyltransferase 6A |
848 |
0.54 |
| chr9_31209643_31210005 | 0.32 |
Aplp2 |
amyloid beta (A4) precursor-like protein 2 |
1963 |
0.34 |
| chr3_107974136_107974298 | 0.32 |
Gstm3 |
glutathione S-transferase, mu 3 |
4934 |
0.08 |
| chr1_187315684_187315899 | 0.32 |
Gm38155 |
predicted gene, 38155 |
61678 |
0.11 |
| chr14_21336208_21336382 | 0.32 |
Adk |
adenosine kinase |
18170 |
0.25 |
| chr5_121334351_121334559 | 0.32 |
Hectd4 |
HECT domain E3 ubiquitin protein ligase 4 |
14432 |
0.12 |
| chr17_5017691_5017888 | 0.32 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
21360 |
0.22 |
| chr5_145858255_145858504 | 0.31 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
18312 |
0.15 |
| chr8_33886292_33886477 | 0.31 |
Gm26978 |
predicted gene, 26978 |
637 |
0.69 |
| chr11_78105085_78105236 | 0.31 |
Fam222b |
family with sequence similarity 222, member B |
10446 |
0.08 |
| chr8_35387028_35387898 | 0.30 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
10803 |
0.16 |
| chr13_34154072_34154234 | 0.30 |
Psmg4 |
proteasome (prosome, macropain) assembly chaperone 4 |
8811 |
0.12 |
| chr2_71254564_71254716 | 0.30 |
Dync1i2 |
dynein cytoplasmic 1 intermediate chain 2 |
5907 |
0.22 |
| chr15_35886635_35886786 | 0.30 |
Vps13b |
vacuolar protein sorting 13B |
14988 |
0.16 |
| chr8_108789308_108789477 | 0.29 |
Gm38042 |
predicted gene, 38042 |
51799 |
0.14 |
| chr15_36335633_36335784 | 0.29 |
Gm33936 |
predicted gene, 33936 |
14046 |
0.12 |
| chr4_11612532_11612699 | 0.29 |
Gm11832 |
predicted gene 11832 |
2392 |
0.23 |
| chr16_93875336_93875598 | 0.29 |
Chaf1b |
chromatin assembly factor 1, subunit B (p60) |
8434 |
0.13 |
| chr4_148060264_148060415 | 0.29 |
Mthfr |
methylenetetrahydrofolate reductase |
7661 |
0.09 |
| chr13_113023428_113023726 | 0.29 |
Cdc20b |
cell division cycle 20B |
11534 |
0.08 |
| chr3_95435996_95436155 | 0.28 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
1556 |
0.21 |
| chr14_73383648_73383821 | 0.28 |
Itm2b |
integral membrane protein 2B |
1464 |
0.43 |
| chr5_28056784_28057184 | 0.28 |
Gm26608 |
predicted gene, 26608 |
1525 |
0.35 |
| chr4_52517213_52517398 | 0.28 |
Vma21-ps |
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae), pseudogene |
20061 |
0.19 |
| chr13_9014068_9014362 | 0.28 |
Gtpbp4 |
GTP binding protein 4 |
18132 |
0.1 |
| chr6_121143927_121144113 | 0.28 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
13021 |
0.14 |
| chr15_73394628_73394911 | 0.28 |
Ptk2 |
PTK2 protein tyrosine kinase 2 |
70 |
0.97 |
| chr2_69336183_69336378 | 0.27 |
Abcb11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
6320 |
0.19 |
| chr1_140173150_140173301 | 0.27 |
Cfh |
complement component factor h |
10055 |
0.24 |
| chr18_32557156_32557525 | 0.27 |
Gypc |
glycophorin C |
2640 |
0.3 |
| chr13_41214741_41215042 | 0.27 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
5271 |
0.14 |
| chr15_100604875_100605219 | 0.27 |
Gm49492 |
predicted gene, 49492 |
4843 |
0.09 |
| chr1_67200520_67200671 | 0.27 |
Gm15668 |
predicted gene 15668 |
48605 |
0.14 |
| chr15_80674781_80675071 | 0.27 |
Fam83f |
family with sequence similarity 83, member F |
3079 |
0.17 |
| chr11_8523561_8523765 | 0.27 |
Tns3 |
tensin 3 |
21688 |
0.27 |
| chr6_147954211_147954549 | 0.27 |
Far2 |
fatty acyl CoA reductase 2 |
92879 |
0.07 |
| chrX_103820422_103820573 | 0.26 |
Slc16a2 |
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
1486 |
0.35 |
| chr1_171216477_171216936 | 0.26 |
Nr1i3 |
nuclear receptor subfamily 1, group I, member 3 |
2357 |
0.12 |
| chr8_40882534_40882732 | 0.26 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
7685 |
0.16 |
| chr4_97789633_97790009 | 0.26 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
11743 |
0.2 |
| chr4_97933124_97933499 | 0.26 |
Nfia |
nuclear factor I/A |
22278 |
0.26 |
| chr11_120674778_120674973 | 0.26 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
1471 |
0.16 |
| chr15_67420672_67420895 | 0.26 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
194014 |
0.03 |
| chr19_55566077_55566254 | 0.26 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
175655 |
0.03 |
| chr13_34735060_34735220 | 0.26 |
Fam50b |
family with sequence similarity 50, member B |
290 |
0.85 |
| chr9_101093226_101093396 | 0.26 |
Gm37553 |
predicted gene, 37553 |
334 |
0.81 |
| chr4_84619745_84620056 | 0.26 |
Bnc2 |
basonuclin 2 |
55096 |
0.15 |
| chr6_128132951_128133168 | 0.25 |
Tspan9 |
tetraspanin 9 |
3650 |
0.16 |
| chr14_89611447_89611598 | 0.25 |
Gm25415 |
predicted gene, 25415 |
95283 |
0.09 |
| chr10_110706552_110706703 | 0.25 |
E2f7 |
E2F transcription factor 7 |
38812 |
0.16 |
| chr10_68103006_68103157 | 0.25 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
33545 |
0.18 |
| chr3_81674937_81675101 | 0.25 |
Gm43346 |
predicted gene 43346 |
46657 |
0.17 |
| chr9_70278042_70278225 | 0.25 |
Myo1e |
myosin IE |
70765 |
0.09 |
| chr6_140440823_140441093 | 0.25 |
Plekha5 |
pleckstrin homology domain containing, family A member 5 |
7969 |
0.22 |
| chr15_76251124_76251290 | 0.25 |
Mir6953 |
microRNA 6953 |
3016 |
0.09 |
| chr2_75668710_75668893 | 0.25 |
Hnrnpa3 |
heterogeneous nuclear ribonucleoprotein A3 |
6312 |
0.13 |
| chr4_145087410_145087580 | 0.25 |
Vps13d |
vacuolar protein sorting 13D |
4455 |
0.3 |
| chr6_72609129_72609280 | 0.25 |
Gm20536 |
predicted gene 20536 |
632 |
0.48 |
| chr18_44785957_44786151 | 0.25 |
Mcc |
mutated in colorectal cancers |
26128 |
0.15 |
| chr6_116064213_116064364 | 0.25 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
6239 |
0.19 |
| chr9_74979549_74979946 | 0.24 |
Fam214a |
family with sequence similarity 214, member A |
3636 |
0.24 |
| chr11_4123188_4123541 | 0.24 |
Sec14l2 |
SEC14-like lipid binding 2 |
51 |
0.94 |
| chr2_6402913_6403256 | 0.24 |
Usp6nl |
USP6 N-terminal like |
17838 |
0.18 |
| chr16_13306126_13306394 | 0.24 |
Mrtfb |
myocardin related transcription factor B |
41184 |
0.16 |
| chr5_25227061_25227222 | 0.24 |
E130116L18Rik |
RIKEN cDNA E130116L18 gene |
3988 |
0.18 |
| chr7_143052216_143052572 | 0.24 |
Cd81 |
CD81 antigen |
345 |
0.72 |
| chr5_44239117_44239268 | 0.24 |
Tapt1 |
transmembrane anterior posterior transformation 1 |
12566 |
0.11 |
| chr10_21448600_21448779 | 0.24 |
Gm48386 |
predicted gene, 48386 |
3053 |
0.2 |
| chr14_51064256_51064407 | 0.24 |
Rnase12 |
ribonuclease, RNase A family, 12 (non-active) |
6670 |
0.08 |
| chr8_95432921_95433173 | 0.24 |
Cfap20 |
cilia and flagella associated protein 20 |
1710 |
0.27 |
| chr4_106331582_106331733 | 0.24 |
Usp24 |
ubiquitin specific peptidase 24 |
15423 |
0.16 |
| chr7_136493461_136493612 | 0.24 |
Gm36849 |
predicted gene, 36849 |
140172 |
0.04 |
| chr17_31872001_31872152 | 0.24 |
Sik1 |
salt inducible kinase 1 |
16272 |
0.13 |
| chr14_70393462_70393631 | 0.24 |
Gm22725 |
predicted gene, 22725 |
28734 |
0.08 |
| chr15_62394159_62394346 | 0.24 |
Pvt1 |
Pvt1 oncogene |
171649 |
0.03 |
| chr3_108770957_108771116 | 0.23 |
Aknad1 |
AKNA domain containing 1 |
10822 |
0.15 |
| chr1_194425856_194426037 | 0.23 |
Plxna2 |
plexin A2 |
192272 |
0.03 |
| chr14_120498503_120498675 | 0.23 |
Rap2a |
RAS related protein 2a |
20145 |
0.24 |
| chr11_116108287_116108624 | 0.23 |
Trim47 |
tripartite motif-containing 47 |
336 |
0.77 |
| chr4_12237364_12237535 | 0.23 |
Gm11847 |
predicted gene 11847 |
4616 |
0.22 |
| chr11_119986483_119986646 | 0.23 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
11107 |
0.11 |
| chr16_37588570_37588721 | 0.23 |
Gm46559 |
predicted gene, 46559 |
7545 |
0.15 |
| chr7_66083686_66083837 | 0.23 |
Selenos |
selenoprotein S |
3800 |
0.14 |
| chr8_33867033_33867186 | 0.23 |
Rbpms |
RNA binding protein gene with multiple splicing |
13436 |
0.16 |
| chr2_103177688_103177876 | 0.23 |
Gm13874 |
predicted gene 13874 |
5 |
0.98 |
| chr4_47366562_47366857 | 0.23 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr7_113778913_113779066 | 0.23 |
Spon1 |
spondin 1, (f-spondin) extracellular matrix protein |
12815 |
0.22 |
| chr6_71837905_71838056 | 0.23 |
Gm44771 |
predicted gene 44771 |
6270 |
0.11 |
| chr11_109926883_109927034 | 0.23 |
Gm11697 |
predicted gene 11697 |
9358 |
0.22 |
| chr13_36533643_36534408 | 0.23 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
3072 |
0.22 |
| chr4_49491887_49492038 | 0.23 |
Baat |
bile acid-Coenzyme A: amino acid N-acyltransferase |
11158 |
0.12 |
| chr13_117744891_117745098 | 0.23 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
24983 |
0.26 |
| chr10_69205623_69205774 | 0.23 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2854 |
0.25 |
| chr4_90495794_90495984 | 0.23 |
Gm12635 |
predicted gene 12635 |
43015 |
0.17 |
| chr9_118044918_118045069 | 0.22 |
Azi2 |
5-azacytidine induced gene 2 |
4306 |
0.22 |
| chr7_127214965_127215278 | 0.22 |
Septin1 |
septin 1 |
4 |
0.92 |
| chr6_42251921_42252091 | 0.22 |
Gstk1 |
glutathione S-transferase kappa 1 |
5983 |
0.11 |
| chr4_6265892_6266043 | 0.22 |
Gm11798 |
predicted gene 11798 |
5002 |
0.21 |
| chr3_10304842_10305071 | 0.22 |
A930014E01Rik |
RIKEN cDNA A930014E01 gene |
165 |
0.9 |
| chr16_58728459_58728625 | 0.22 |
Cldnd1 |
claudin domain containing 1 |
494 |
0.67 |
| chr11_104539702_104539853 | 0.22 |
Cdc27 |
cell division cycle 27 |
10615 |
0.11 |
| chr7_142533086_142533550 | 0.22 |
Mrpl23 |
mitochondrial ribosomal protein L23 |
156 |
0.91 |
| chr4_99046271_99046475 | 0.22 |
Dock7 |
dedicator of cytokinesis 7 |
8960 |
0.18 |
| chr13_34650669_34650865 | 0.22 |
Pxdc1 |
PX domain containing 1 |
1914 |
0.22 |
| chr1_88006329_88006480 | 0.22 |
Usp40 |
ubiquitin specific peptidase 40 |
1825 |
0.18 |
| chr1_30941324_30941475 | 0.22 |
Ptp4a1 |
protein tyrosine phosphatase 4a1 |
2905 |
0.22 |
| chr2_103598963_103599142 | 0.21 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
32742 |
0.16 |
| chr14_21033435_21033611 | 0.21 |
Vcl |
vinculin |
11520 |
0.18 |
| chr5_45430764_45431089 | 0.21 |
Gm42413 |
predicted gene, 42413 |
3463 |
0.17 |
| chr10_40297176_40297522 | 0.21 |
Amd1 |
S-adenosylmethionine decarboxylase 1 |
4227 |
0.13 |
| chr2_35103269_35103589 | 0.21 |
AI182371 |
expressed sequence AI182371 |
1886 |
0.29 |
| chr3_81997803_81997954 | 0.21 |
Asic5 |
acid-sensing (proton-gated) ion channel family member 5 |
956 |
0.53 |
| chr9_57439182_57439333 | 0.21 |
Ppcdc |
phosphopantothenoylcysteine decarboxylase |
834 |
0.49 |
| chr14_17696257_17696429 | 0.21 |
Thrb |
thyroid hormone receptor beta |
35447 |
0.21 |
| chr1_74901365_74901516 | 0.21 |
Mir375 |
microRNA 375 |
719 |
0.39 |
| chr2_67881464_67881701 | 0.21 |
Gm37964 |
predicted gene, 37964 |
17014 |
0.24 |
| chr13_90359772_90359986 | 0.21 |
Gm37708 |
predicted gene, 37708 |
26678 |
0.21 |
| chr10_81091333_81091491 | 0.21 |
Creb3l3 |
cAMP responsive element binding protein 3-like 3 |
494 |
0.58 |
| chr4_132997648_132997799 | 0.21 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
13537 |
0.16 |
| chr7_102293168_102293449 | 0.21 |
Stim1 |
stromal interaction molecule 1 |
24981 |
0.12 |
| chr5_115018762_115018913 | 0.21 |
Sppl3 |
signal peptide peptidase 3 |
7340 |
0.1 |
| chr6_127108825_127109062 | 0.21 |
Tigar |
Trp53 induced glycolysis regulatory phosphatase |
607 |
0.6 |
| chr4_101282048_101282334 | 0.21 |
Gm12801 |
predicted gene 12801 |
2030 |
0.22 |
| chr2_31484723_31485077 | 0.21 |
Ass1 |
argininosuccinate synthetase 1 |
12872 |
0.18 |
| chr12_30341867_30342052 | 0.21 |
Sntg2 |
syntrophin, gamma 2 |
15724 |
0.25 |
| chr19_32807644_32807795 | 0.21 |
Pten |
phosphatase and tensin homolog |
3506 |
0.32 |
| chr15_62112429_62112580 | 0.21 |
Pvt1 |
Pvt1 oncogene |
13475 |
0.27 |
| chr6_145793553_145793704 | 0.21 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
14755 |
0.19 |
| chr10_31650590_31650759 | 0.21 |
Gm8793 |
predicted gene 8793 |
11516 |
0.21 |
| chr17_83181717_83182272 | 0.20 |
Pkdcc |
protein kinase domain containing, cytoplasmic |
33298 |
0.17 |
| chr12_34238298_34238489 | 0.20 |
Gm18025 |
predicted gene, 18025 |
52742 |
0.18 |
| chr11_73048783_73048934 | 0.20 |
Ncbp3 |
nuclear cap binding subunit 3 |
985 |
0.43 |
| chr3_135675431_135675674 | 0.20 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
6213 |
0.21 |
| chr16_95139892_95140046 | 0.20 |
Gm49642 |
predicted gene, 49642 |
31541 |
0.18 |
| chr5_72883801_72883981 | 0.20 |
Tec |
tec protein tyrosine kinase |
15408 |
0.14 |
| chr2_178680469_178680632 | 0.20 |
Cdh26 |
cadherin-like 26 |
219920 |
0.02 |
| chr12_102271886_102272090 | 0.20 |
Rin3 |
Ras and Rab interactor 3 |
11060 |
0.21 |
| chr11_78103949_78104116 | 0.20 |
Fam222b |
family with sequence similarity 222, member B |
9324 |
0.08 |
| chr6_139875525_139875713 | 0.20 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
31971 |
0.17 |
| chr11_34834139_34834311 | 0.20 |
Spdl1 |
spindle apparatus coiled-coil protein 1 |
584 |
0.74 |
| chr17_50544760_50544923 | 0.20 |
Plcl2 |
phospholipase C-like 2 |
35438 |
0.22 |
| chr16_33222596_33222759 | 0.20 |
Osbpl11 |
oxysterol binding protein-like 11 |
1921 |
0.33 |
| chr15_84412819_84412970 | 0.19 |
Shisal1 |
shisa like 1 |
30803 |
0.16 |
| chr1_21262754_21263025 | 0.19 |
Gm28836 |
predicted gene 28836 |
8704 |
0.11 |
| chr10_127082531_127082956 | 0.19 |
Agap2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
3836 |
0.09 |
| chr2_75722764_75722925 | 0.19 |
Gm13656 |
predicted gene 13656 |
7328 |
0.13 |
| chr1_151325474_151325655 | 0.19 |
Gm10138 |
predicted gene 10138 |
18518 |
0.13 |
| chr8_44971130_44971287 | 0.19 |
Fat1 |
FAT atypical cadherin 1 |
20994 |
0.18 |
| chr10_84885783_84886161 | 0.19 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31644 |
0.17 |
| chr15_80680833_80681008 | 0.19 |
Fam83f |
family with sequence similarity 83, member F |
9073 |
0.13 |
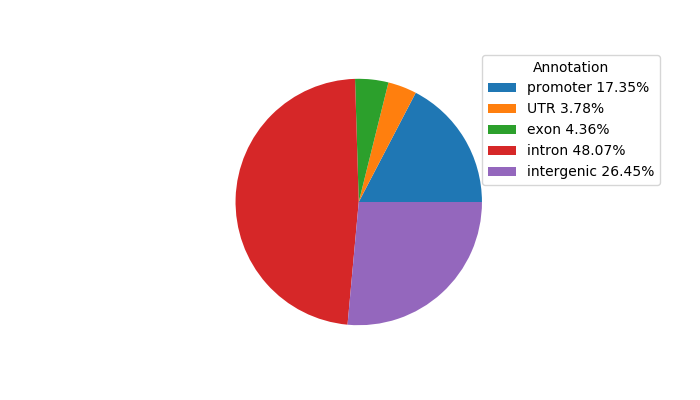
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.1 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.0 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |