Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
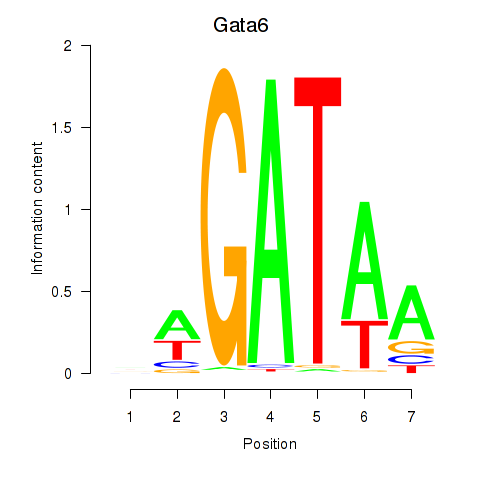
Results for Gata6
Z-value: 1.61
Transcription factors associated with Gata6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata6
|
ENSMUSG00000005836.8 | GATA binding protein 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_11209102_11209260 | Gata6 | 150134 | 0.040384 | -0.93 | 7.1e-03 | Click! |
| chr18_11052288_11052439 | Gata6 | 107 | 0.589768 | -0.80 | 5.8e-02 | Click! |
| chr18_11210913_11211071 | Gata6 | 151945 | 0.039601 | -0.76 | 8.1e-02 | Click! |
| chr18_11221965_11222116 | Gata6 | 162993 | 0.035259 | -0.71 | 1.1e-01 | Click! |
| chr18_11271924_11272075 | Gata6 | 212952 | 0.022153 | -0.70 | 1.2e-01 | Click! |
Activity of the Gata6 motif across conditions
Conditions sorted by the z-value of the Gata6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
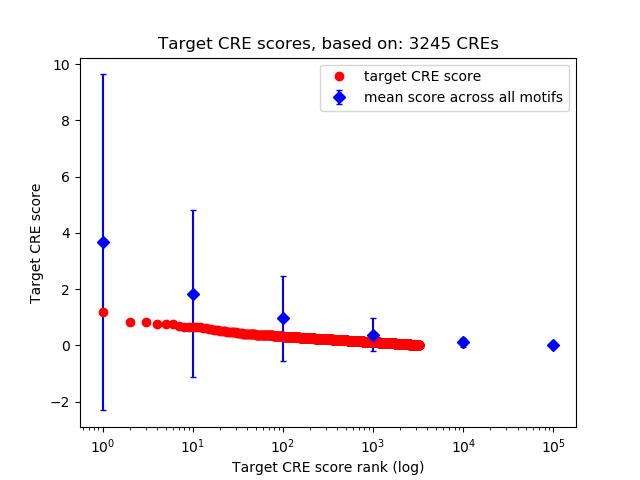
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_44437957_44438156 | 1.17 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
1656 |
0.27 |
| chr3_138288275_138288480 | 0.84 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
10726 |
0.12 |
| chr9_74892080_74892501 | 0.83 |
Onecut1 |
one cut domain, family member 1 |
25806 |
0.14 |
| chr19_40146515_40146666 | 0.77 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
40696 |
0.11 |
| chr3_89149636_89149787 | 0.76 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
1503 |
0.17 |
| chr7_97414019_97414355 | 0.74 |
Thrsp |
thyroid hormone responsive |
3332 |
0.16 |
| chr9_74523907_74524058 | 0.70 |
Gm28622 |
predicted gene 28622 |
37396 |
0.17 |
| chr6_149169218_149169449 | 0.65 |
Amn1 |
antagonist of mitotic exit network 1 |
4038 |
0.17 |
| chr1_162986568_162986732 | 0.64 |
Fmo3 |
flavin containing monooxygenase 3 |
2122 |
0.27 |
| chr1_21248546_21248733 | 0.64 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4882 |
0.13 |
| chr8_93193292_93193577 | 0.64 |
Gm45909 |
predicted gene 45909 |
2076 |
0.23 |
| chr9_74891211_74891440 | 0.63 |
Onecut1 |
one cut domain, family member 1 |
24841 |
0.14 |
| chr11_120815283_120815743 | 0.62 |
Fasn |
fatty acid synthase |
58 |
0.94 |
| chr4_141955373_141955574 | 0.61 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1279 |
0.36 |
| chr18_33351542_33351716 | 0.57 |
Gm5503 |
predicted gene 5503 |
33326 |
0.21 |
| chr3_18178649_18178800 | 0.57 |
Gm23686 |
predicted gene, 23686 |
1099 |
0.59 |
| chr8_105095555_105095947 | 0.54 |
Ces3b |
carboxylesterase 3B |
7132 |
0.11 |
| chr8_44373378_44373540 | 0.53 |
Gm37972 |
predicted gene, 37972 |
144914 |
0.04 |
| chr10_110717608_110717759 | 0.53 |
E2f7 |
E2F transcription factor 7 |
27756 |
0.19 |
| chr2_155102805_155102956 | 0.53 |
2310005A03Rik |
RIKEN cDNA 2310005A03 gene |
2799 |
0.2 |
| chr10_87917557_87917708 | 0.50 |
Tyms-ps |
thymidylate synthase, pseudogene |
49215 |
0.11 |
| chr11_90321650_90321810 | 0.50 |
Hlf |
hepatic leukemia factor |
60205 |
0.12 |
| chr6_127045144_127045338 | 0.49 |
Fgf23 |
fibroblast growth factor 23 |
27661 |
0.1 |
| chr15_7175278_7175429 | 0.48 |
Lifr |
LIF receptor alpha |
21000 |
0.23 |
| chr6_51695515_51695666 | 0.48 |
Gm38811 |
predicted gene, 38811 |
15491 |
0.23 |
| chr8_105095287_105095438 | 0.48 |
Ces3b |
carboxylesterase 3B |
6743 |
0.11 |
| chr11_16823614_16823889 | 0.47 |
Egfros |
epidermal growth factor receptor, opposite strand |
6951 |
0.23 |
| chr6_22037743_22037894 | 0.47 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
11875 |
0.27 |
| chr3_148965860_148966026 | 0.47 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
11156 |
0.2 |
| chr9_107307510_107307734 | 0.46 |
Gm17041 |
predicted gene 17041 |
5784 |
0.1 |
| chr11_16854059_16854210 | 0.45 |
Egfros |
epidermal growth factor receptor, opposite strand |
23432 |
0.17 |
| chr5_144364197_144364385 | 0.45 |
Dmrt1i |
Dmrt1 interacting ncRNA |
5766 |
0.18 |
| chr3_133740753_133740929 | 0.44 |
Gm6135 |
prediticted gene 6135 |
50663 |
0.13 |
| chr8_36242947_36243098 | 0.44 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
6494 |
0.21 |
| chr15_3640425_3640638 | 0.43 |
Gm22031 |
predicted gene, 22031 |
37134 |
0.18 |
| chr6_121171179_121171645 | 0.42 |
Pex26 |
peroxisomal biogenesis factor 26 |
12255 |
0.13 |
| chr1_67230512_67230663 | 0.42 |
Gm15668 |
predicted gene 15668 |
18613 |
0.21 |
| chr10_95434881_95435051 | 0.41 |
5730420D15Rik |
RIKEN cDNA 5730420D15 gene |
17591 |
0.11 |
| chr11_16838643_16838829 | 0.41 |
Egfros |
epidermal growth factor receptor, opposite strand |
8034 |
0.22 |
| chr4_45403274_45403659 | 0.41 |
Slc25a51 |
solute carrier family 25, member 51 |
1400 |
0.35 |
| chr4_129063899_129064050 | 0.40 |
Rnf19b |
ring finger protein 19B |
5160 |
0.15 |
| chr11_16862611_16863177 | 0.40 |
Egfr |
epidermal growth factor receptor |
15256 |
0.19 |
| chr1_162892544_162892705 | 0.40 |
Fmo2 |
flavin containing monooxygenase 2 |
5860 |
0.19 |
| chr9_122634913_122635137 | 0.40 |
Gm47134 |
predicted gene, 47134 |
12805 |
0.13 |
| chr4_76963069_76963220 | 0.40 |
Gm23159 |
predicted gene, 23159 |
4646 |
0.28 |
| chr12_40574749_40574900 | 0.40 |
Dock4 |
dedicator of cytokinesis 4 |
128488 |
0.05 |
| chr8_38604147_38604298 | 0.39 |
Gm40493 |
predicted gene, 40493 |
55541 |
0.14 |
| chrX_156419717_156419882 | 0.39 |
Gm23404 |
predicted gene, 23404 |
36296 |
0.23 |
| chr3_18180658_18180809 | 0.39 |
Gm23686 |
predicted gene, 23686 |
3108 |
0.31 |
| chr2_58765903_58766054 | 0.38 |
Upp2 |
uridine phosphorylase 2 |
653 |
0.74 |
| chr15_59195501_59195712 | 0.38 |
Rpl7-ps8 |
ribosomal protein L7, pseudogene 8 |
15302 |
0.19 |
| chr6_85810872_85811030 | 0.38 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
1088 |
0.31 |
| chr9_74326904_74327074 | 0.38 |
Gm24141 |
predicted gene, 24141 |
35621 |
0.16 |
| chr1_130733366_130733529 | 0.38 |
AA986860 |
expressed sequence AA986860 |
1337 |
0.24 |
| chr5_90475088_90475243 | 0.38 |
Alb |
albumin |
12465 |
0.14 |
| chr6_66996154_66996403 | 0.37 |
Gm36816 |
predicted gene, 36816 |
12135 |
0.12 |
| chr12_104391135_104391367 | 0.37 |
Serpina3m |
serine (or cysteine) peptidase inhibitor, clade A, member 3M |
4087 |
0.15 |
| chr3_18111157_18111308 | 0.37 |
Gm23726 |
predicted gene, 23726 |
41906 |
0.14 |
| chr13_82200374_82200525 | 0.37 |
Gm48155 |
predicted gene, 48155 |
110692 |
0.07 |
| chr12_32681759_32681933 | 0.37 |
Gm47937 |
predicted gene, 47937 |
8552 |
0.23 |
| chr6_142471603_142471968 | 0.37 |
Gys2 |
glycogen synthase 2 |
1324 |
0.42 |
| chrY_90794856_90795007 | 0.37 |
Gm47283 |
predicted gene, 47283 |
4480 |
0.21 |
| chr1_153065706_153066077 | 0.37 |
Gm28960 |
predicted gene 28960 |
23616 |
0.16 |
| chr15_59044505_59044656 | 0.36 |
Mtss1 |
MTSS I-BAR domain containing 1 |
3983 |
0.26 |
| chr1_67223699_67224148 | 0.36 |
Gm15668 |
predicted gene 15668 |
25277 |
0.19 |
| chr9_74695756_74695907 | 0.36 |
Gm27233 |
predicted gene 27233 |
13431 |
0.23 |
| chr5_121947991_121948177 | 0.36 |
Cux2 |
cut-like homeobox 2 |
23163 |
0.15 |
| chr7_120028626_120028778 | 0.36 |
Gm25217 |
predicted gene, 25217 |
58881 |
0.08 |
| chr9_100535718_100535869 | 0.36 |
Gm28586 |
predicted gene 28586 |
4722 |
0.16 |
| chr11_16859851_16860002 | 0.35 |
Egfr |
epidermal growth factor receptor |
18224 |
0.18 |
| chr9_120133572_120133723 | 0.35 |
Gm5922 |
predicted gene 5922 |
1368 |
0.2 |
| chr13_112123093_112123287 | 0.35 |
Gm31104 |
predicted gene, 31104 |
14926 |
0.2 |
| chr11_16837780_16838032 | 0.35 |
Egfros |
epidermal growth factor receptor, opposite strand |
7204 |
0.22 |
| chr3_18167606_18167757 | 0.35 |
Gm23686 |
predicted gene, 23686 |
9944 |
0.24 |
| chr8_114154739_114154891 | 0.35 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
21173 |
0.24 |
| chr4_104908553_104908715 | 0.35 |
Fyb2 |
FYN binding protein 2 |
4822 |
0.23 |
| chr8_77477761_77477925 | 0.35 |
0610038B21Rik |
RIKEN cDNA 0610038B21 gene |
39213 |
0.12 |
| chr4_102586727_102587163 | 0.35 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
613 |
0.84 |
| chr19_10065348_10065653 | 0.35 |
Fads2 |
fatty acid desaturase 2 |
5687 |
0.14 |
| chr7_98350774_98350925 | 0.35 |
Tsku |
tsukushi, small leucine rich proteoglycan |
9230 |
0.17 |
| chr4_102576883_102577034 | 0.34 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
6863 |
0.31 |
| chr2_85351619_85351794 | 0.34 |
Olfr988 |
olfactory receptor 988 |
2294 |
0.18 |
| chr7_114362001_114362175 | 0.33 |
4933406I18Rik |
RIKEN cDNA 4933406I18 gene |
52933 |
0.12 |
| chr8_115830386_115830552 | 0.33 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
122675 |
0.06 |
| chr11_16781109_16781403 | 0.33 |
Egfr |
epidermal growth factor receptor |
29026 |
0.16 |
| chr11_118266078_118266308 | 0.33 |
Usp36 |
ubiquitin specific peptidase 36 |
145 |
0.95 |
| chr14_46615680_46615835 | 0.33 |
Gm49319 |
predicted gene, 49319 |
4747 |
0.13 |
| chr2_52613475_52613626 | 0.33 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
6204 |
0.24 |
| chr15_3469696_3469847 | 0.33 |
Ghr |
growth hormone receptor |
1873 |
0.48 |
| chr1_130767394_130767576 | 0.33 |
Gm28856 |
predicted gene 28856 |
1101 |
0.34 |
| chr17_47304805_47304956 | 0.33 |
Trerf1 |
transcriptional regulating factor 1 |
10811 |
0.16 |
| chr5_106224440_106224591 | 0.33 |
Gm5987 |
predicted gene 5987 |
8864 |
0.21 |
| chr14_21261451_21261651 | 0.33 |
Adk |
adenosine kinase |
56574 |
0.15 |
| chr15_81847424_81847575 | 0.32 |
Gm8444 |
predicted gene 8444 |
3786 |
0.11 |
| chr15_58970873_58971171 | 0.32 |
Mtss1 |
MTSS I-BAR domain containing 1 |
1517 |
0.34 |
| chr19_10094218_10094369 | 0.32 |
Fads2 |
fatty acid desaturase 2 |
5577 |
0.16 |
| chr2_71874267_71874418 | 0.32 |
Pdk1 |
pyruvate dehydrogenase kinase, isoenzyme 1 |
599 |
0.75 |
| chr5_90475599_90475916 | 0.32 |
Alb |
albumin |
13057 |
0.14 |
| chr15_3470264_3470480 | 0.32 |
Ghr |
growth hormone receptor |
1272 |
0.6 |
| chr19_18128513_18128741 | 0.32 |
Gm18610 |
predicted gene, 18610 |
77168 |
0.1 |
| chr5_125519617_125519768 | 0.31 |
Aacs |
acetoacetyl-CoA synthetase |
4449 |
0.18 |
| chr8_93126283_93126451 | 0.31 |
Ces1c |
carboxylesterase 1C |
1838 |
0.28 |
| chr15_53199808_53200033 | 0.31 |
Ext1 |
exostosin glycosyltransferase 1 |
3665 |
0.38 |
| chr3_149181265_149181842 | 0.31 |
Gm42647 |
predicted gene 42647 |
28718 |
0.19 |
| chr11_84243201_84243352 | 0.31 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
37179 |
0.15 |
| chr11_16893716_16893915 | 0.31 |
Egfr |
epidermal growth factor receptor |
11370 |
0.19 |
| chr8_93236044_93236211 | 0.31 |
Ces1e |
carboxylesterase 1E |
6508 |
0.14 |
| chr13_4525310_4525461 | 0.31 |
Akr1c20 |
aldo-keto reductase family 1, member C20 |
2040 |
0.31 |
| chr10_69206165_69206505 | 0.31 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2217 |
0.29 |
| chr10_87867374_87867656 | 0.31 |
Igf1os |
insulin-like growth factor 1, opposite strand |
4134 |
0.23 |
| chr11_51739157_51739308 | 0.31 |
Gm25291 |
predicted gene, 25291 |
1310 |
0.37 |
| chr1_70927541_70927750 | 0.31 |
Gm16236 |
predicted gene 16236 |
112385 |
0.07 |
| chr16_77048481_77048632 | 0.31 |
Usp25 |
ubiquitin specific peptidase 25 |
32946 |
0.18 |
| chr2_122251002_122251156 | 0.31 |
Sord |
sorbitol dehydrogenase |
16330 |
0.1 |
| chr6_17185802_17185953 | 0.31 |
D830026I12Rik |
RIKEN cDNA D830026I12 gene |
11874 |
0.2 |
| chr17_81174915_81175073 | 0.30 |
Gm50042 |
predicted gene, 50042 |
71744 |
0.11 |
| chr3_60965282_60965682 | 0.30 |
P2ry1 |
purinergic receptor P2Y, G-protein coupled 1 |
37313 |
0.16 |
| chr16_45366538_45366697 | 0.30 |
Cd200 |
CD200 antigen |
33695 |
0.14 |
| chr17_78392816_78393001 | 0.30 |
Fez2 |
fasciculation and elongation protein zeta 2 (zygin II) |
8522 |
0.17 |
| chr12_104340629_104340859 | 0.30 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
2258 |
0.18 |
| chr3_117078252_117078413 | 0.30 |
1700061I17Rik |
RIKEN cDNA 1700061I17 gene |
567 |
0.75 |
| chr4_82338577_82338767 | 0.30 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
100738 |
0.08 |
| chr4_150379881_150380059 | 0.30 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
26424 |
0.17 |
| chr13_62862796_62863198 | 0.30 |
Fbp2 |
fructose bisphosphatase 2 |
4575 |
0.14 |
| chr2_31483571_31483901 | 0.30 |
Ass1 |
argininosuccinate synthetase 1 |
13529 |
0.18 |
| chrX_98052908_98053108 | 0.30 |
Gm14802 |
predicted gene 14802 |
12633 |
0.29 |
| chr14_75054836_75055035 | 0.29 |
Rubcnl |
RUN and cysteine rich domain containing beclin 1 interacting protein like |
18313 |
0.18 |
| chr1_67211421_67211668 | 0.29 |
Gm15668 |
predicted gene 15668 |
37656 |
0.16 |
| chr7_101085910_101086353 | 0.29 |
Fchsd2 |
FCH and double SH3 domains 2 |
6732 |
0.17 |
| chr19_3556269_3556436 | 0.29 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
19344 |
0.14 |
| chr12_25861580_25861731 | 0.29 |
Gm47733 |
predicted gene, 47733 |
9498 |
0.26 |
| chr2_72182811_72183066 | 0.29 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
3245 |
0.25 |
| chr16_72639482_72639633 | 0.29 |
Robo1 |
roundabout guidance receptor 1 |
23592 |
0.28 |
| chr10_125282815_125282979 | 0.29 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
25919 |
0.2 |
| chr6_85763479_85763641 | 0.29 |
Nat8f3 |
N-acetyltransferase 8 (GCN5-related) family member 3 |
1080 |
0.33 |
| chr1_21306552_21306732 | 0.29 |
Gm4956 |
predicted gene 4956 |
8330 |
0.1 |
| chr12_78269881_78270053 | 0.29 |
Gm48225 |
predicted gene, 48225 |
8674 |
0.17 |
| chr11_78182563_78182714 | 0.29 |
Rpl23a |
ribosomal protein L23A |
439 |
0.34 |
| chr7_114361745_114361949 | 0.29 |
4933406I18Rik |
RIKEN cDNA 4933406I18 gene |
53174 |
0.12 |
| chr14_118948462_118948868 | 0.29 |
Dnajc3 |
DnaJ heat shock protein family (Hsp40) member C3 |
10689 |
0.16 |
| chr7_63899068_63899509 | 0.29 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr4_101717989_101718160 | 0.29 |
Lepr |
leptin receptor |
667 |
0.76 |
| chr11_16836119_16836554 | 0.29 |
Egfros |
epidermal growth factor receptor, opposite strand |
5634 |
0.23 |
| chrY_90802780_90802931 | 0.28 |
Gm47283 |
predicted gene, 47283 |
12404 |
0.17 |
| chr2_155066379_155066530 | 0.28 |
Gm45609 |
predicted gene 45609 |
7727 |
0.14 |
| chr1_70928023_70928290 | 0.28 |
Gm16236 |
predicted gene 16236 |
111874 |
0.07 |
| chr3_136048376_136048532 | 0.28 |
Gm5281 |
predicted gene 5281 |
3234 |
0.26 |
| chr19_21624664_21624891 | 0.28 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
5991 |
0.23 |
| chr2_52576390_52576567 | 0.28 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
17911 |
0.18 |
| chr17_84733210_84733384 | 0.28 |
Lrpprc |
leucine-rich PPR-motif containing |
2113 |
0.28 |
| chr2_3700010_3700171 | 0.28 |
Fam107b |
family with sequence similarity 107, member B |
3923 |
0.2 |
| chr14_17728323_17728474 | 0.28 |
Gm48320 |
predicted gene, 48320 |
42724 |
0.19 |
| chr15_10267200_10267351 | 0.28 |
Prlr |
prolactin receptor |
17715 |
0.23 |
| chr5_123734312_123734463 | 0.28 |
Rsrc2 |
arginine/serine-rich coiled-coil 2 |
693 |
0.57 |
| chr9_64714462_64714631 | 0.28 |
Rab11a |
RAB11A, member RAS oncogene family |
11174 |
0.17 |
| chr3_152347303_152347462 | 0.28 |
Usp33 |
ubiquitin specific peptidase 33 |
867 |
0.48 |
| chr11_11962403_11962585 | 0.28 |
Grb10 |
growth factor receptor bound protein 10 |
6470 |
0.23 |
| chr15_59050746_59050916 | 0.28 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4633 |
0.25 |
| chr9_105792087_105792238 | 0.27 |
Col6a6 |
collagen, type VI, alpha 6 |
17486 |
0.2 |
| chr16_43303061_43303278 | 0.27 |
Gm37946 |
predicted gene, 37946 |
5112 |
0.21 |
| chr12_118313299_118313450 | 0.27 |
Sp4 |
trans-acting transcription factor 4 |
11934 |
0.27 |
| chr3_129444095_129444246 | 0.27 |
Gm5712 |
predicted gene 5712 |
10497 |
0.17 |
| chr11_98289682_98289855 | 0.27 |
Gm20644 |
predicted gene 20644 |
4386 |
0.11 |
| chr18_33377697_33377885 | 0.27 |
Gm5503 |
predicted gene 5503 |
7164 |
0.27 |
| chr2_109721800_109721977 | 0.27 |
Bdnf |
brain derived neurotrophic factor |
12505 |
0.2 |
| chr6_149135432_149135583 | 0.27 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
4342 |
0.14 |
| chr13_44640104_44640258 | 0.27 |
Gm47805 |
predicted gene, 47805 |
17999 |
0.21 |
| chr8_125346624_125346799 | 0.27 |
Gm16237 |
predicted gene 16237 |
119388 |
0.06 |
| chr17_5324481_5324860 | 0.27 |
Gm29050 |
predicted gene 29050 |
64093 |
0.11 |
| chr4_6242659_6242849 | 0.27 |
Gm11798 |
predicted gene 11798 |
18211 |
0.19 |
| chr4_148644834_148645010 | 0.27 |
Gm572 |
predicted gene 572 |
1605 |
0.29 |
| chr3_32563174_32563674 | 0.27 |
Mfn1 |
mitofusin 1 |
516 |
0.74 |
| chrX_170020226_170020377 | 0.27 |
Erdr1 |
erythroid differentiation regulator 1 |
9557 |
0.18 |
| chr7_26816390_26816587 | 0.27 |
Cyp2g1 |
cytochrome P450, family 2, subfamily g, polypeptide 1 |
193 |
0.93 |
| chr17_32946227_32946412 | 0.27 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
1016 |
0.36 |
| chr1_67143312_67143463 | 0.27 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
20361 |
0.22 |
| chr5_125467628_125467779 | 0.27 |
Gm43756 |
predicted gene 43756 |
3794 |
0.14 |
| chr15_58984870_58985094 | 0.27 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
846 |
0.57 |
| chr2_71607065_71607232 | 0.27 |
Dlx2 |
distal-less homeobox 2 |
60394 |
0.09 |
| chr5_90468289_90468670 | 0.27 |
Alb |
albumin |
5779 |
0.16 |
| chr13_101849279_101849430 | 0.27 |
Gm47007 |
predicted gene, 47007 |
6137 |
0.21 |
| chr13_37601794_37602225 | 0.26 |
Gm47753 |
predicted gene, 47753 |
19738 |
0.1 |
| chr4_148610779_148610962 | 0.26 |
Tardbp |
TAR DNA binding protein |
5268 |
0.11 |
| chr7_19756083_19756234 | 0.26 |
Bcam |
basal cell adhesion molecule |
793 |
0.37 |
| chr7_26831798_26832173 | 0.26 |
Cyp2a5 |
cytochrome P450, family 2, subfamily a, polypeptide 5 |
3320 |
0.21 |
| chr3_18246603_18246795 | 0.26 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
3361 |
0.3 |
| chr12_102327592_102327745 | 0.26 |
Rin3 |
Ras and Rab interactor 3 |
27077 |
0.18 |
| chr6_25289113_25289264 | 0.26 |
Gm22529 |
predicted gene, 22529 |
89137 |
0.1 |
| chr4_123990670_123991070 | 0.26 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr17_31324627_31325122 | 0.26 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
753 |
0.58 |
| chr3_149437041_149437628 | 0.26 |
Gm30382 |
predicted gene, 30382 |
7942 |
0.29 |
| chr18_4996636_4996787 | 0.26 |
Svil |
supervillin |
1775 |
0.5 |
| chr9_70006377_70006528 | 0.26 |
Bnip2 |
BCL2/adenovirus E1B interacting protein 2 |
3073 |
0.19 |
| chr1_190057217_190057407 | 0.26 |
Gm28172 |
predicted gene 28172 |
111358 |
0.05 |
| chr12_57538162_57538324 | 0.26 |
Foxa1 |
forkhead box A1 |
7878 |
0.15 |
| chr19_56534525_56534676 | 0.26 |
Dclre1a |
DNA cross-link repair 1A |
2782 |
0.26 |
| chr1_91104292_91104494 | 0.26 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
6821 |
0.19 |
| chr5_90471894_90472247 | 0.26 |
Alb |
albumin |
9370 |
0.15 |
| chr3_130999761_130999912 | 0.26 |
A430072C10Rik |
RIKEN cDNA A430072C10 gene |
26534 |
0.14 |
| chr14_69411185_69411411 | 0.26 |
Gm38378 |
predicted gene, 38378 |
13527 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.0 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:0014894 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:1903301 | positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.0 | GO:0034091 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.0 | GO:0043174 | nucleoside salvage(GO:0043174) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 1.0 | GO:0010296 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.4 | GO:0015932 | nucleobase-containing compound transmembrane transporter activity(GO:0015932) |
| 0.0 | 0.1 | GO:0034819 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |