Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
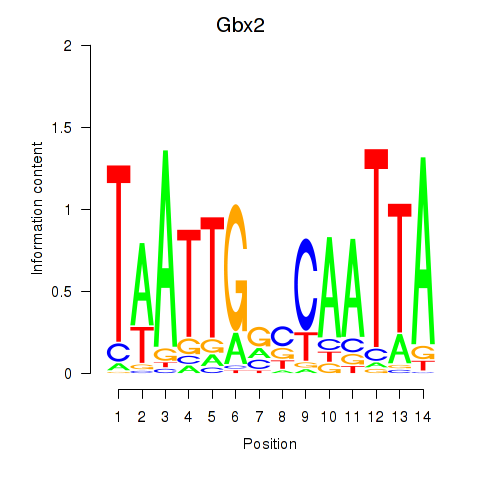
Results for Gbx2
Z-value: 0.60
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSMUSG00000034486.7 | gastrulation brain homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_89940488_89940663 | Gbx2 | 9396 | 0.194843 | 0.71 | 1.2e-01 | Click! |
| chr1_89943333_89943497 | Gbx2 | 12236 | 0.188652 | 0.50 | 3.1e-01 | Click! |
| chr1_89919308_89919468 | Gbx2 | 9797 | 0.198152 | -0.46 | 3.5e-01 | Click! |
| chr1_89895936_89896118 | Gbx2 | 33158 | 0.154999 | -0.32 | 5.3e-01 | Click! |
| chr1_89898377_89898561 | Gbx2 | 30716 | 0.160247 | 0.32 | 5.4e-01 | Click! |
Activity of the Gbx2 motif across conditions
Conditions sorted by the z-value of the Gbx2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
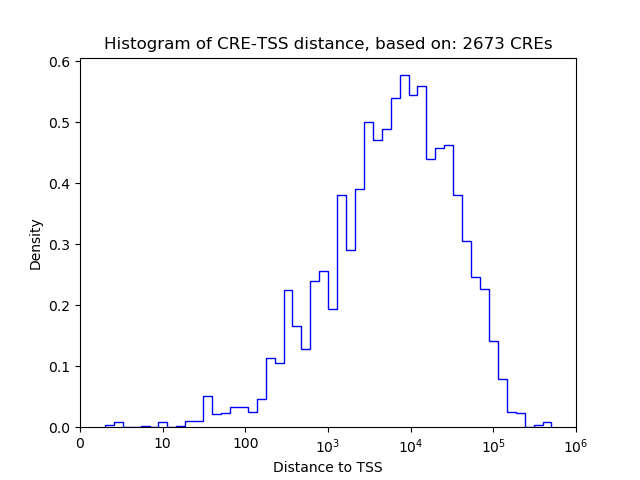
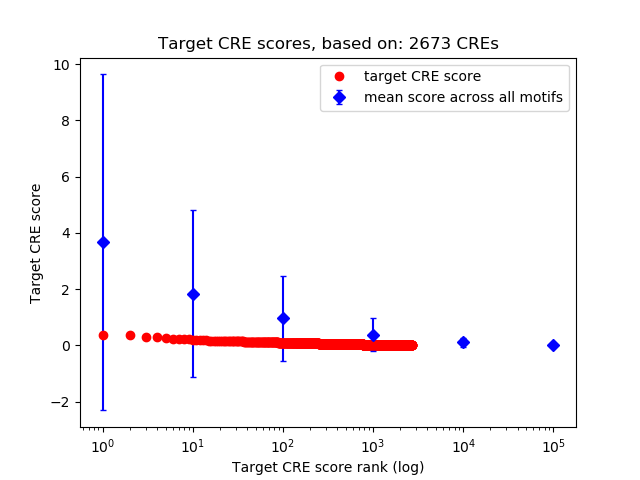
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_24229585_24229736 | 0.37 |
Gm31814 |
predicted gene, 31814 |
13136 |
0.21 |
| chr13_117745874_117746186 | 0.35 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr11_112218841_112219019 | 0.31 |
Gm11680 |
predicted gene 11680 |
111492 |
0.07 |
| chr15_52378702_52379046 | 0.29 |
Gm41322 |
predicted gene, 41322 |
66248 |
0.1 |
| chrX_113094960_113095111 | 0.27 |
Mir361 |
microRNA 361 |
20142 |
0.18 |
| chr5_126536904_126537081 | 0.23 |
Gm24839 |
predicted gene, 24839 |
23011 |
0.2 |
| chr12_82855785_82855951 | 0.23 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
83287 |
0.09 |
| chr5_8979135_8979286 | 0.21 |
Crot |
carnitine O-octanoyltransferase |
2934 |
0.17 |
| chr8_62229066_62229232 | 0.21 |
Gm2961 |
predicted gene 2961 |
34937 |
0.21 |
| chr8_70186123_70186294 | 0.21 |
Tmem161a |
transmembrane protein 161A |
5669 |
0.1 |
| chr2_88008140_88008468 | 0.20 |
Olfr1160 |
olfactory receptor 1160 |
36 |
0.94 |
| chr8_117737291_117737442 | 0.18 |
Gm31774 |
predicted gene, 31774 |
10894 |
0.14 |
| chr15_36578902_36579081 | 0.18 |
Gm44310 |
predicted gene, 44310 |
4553 |
0.16 |
| chr18_44633864_44634028 | 0.17 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
27719 |
0.19 |
| chr19_30093178_30093359 | 0.17 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
1307 |
0.48 |
| chr19_5073247_5073404 | 0.17 |
Gm46658 |
predicted gene, 46658 |
1096 |
0.2 |
| chr7_113230366_113230557 | 0.17 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
4162 |
0.25 |
| chr4_69993246_69993404 | 0.17 |
Gm11224 |
predicted gene 11224 |
38049 |
0.17 |
| chr11_16753728_16753924 | 0.16 |
Egfr |
epidermal growth factor receptor |
1596 |
0.4 |
| chr19_38358035_38358349 | 0.16 |
Gm50150 |
predicted gene, 50150 |
15578 |
0.13 |
| chr16_84807428_84807749 | 0.16 |
Jam2 |
junction adhesion molecule 2 |
1819 |
0.26 |
| chr6_93153422_93153582 | 0.15 |
Gm5313 |
predicted gene 5313 |
15512 |
0.21 |
| chr5_151104950_151105162 | 0.15 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
3679 |
0.31 |
| chr19_34852544_34852695 | 0.15 |
Pank1 |
pantothenate kinase 1 |
25230 |
0.13 |
| chr7_143068403_143068570 | 0.15 |
Tssc4 |
tumor-suppressing subchromosomal transferable fragment 4 |
763 |
0.46 |
| chr12_44222413_44222596 | 0.15 |
Gm2027 |
predicted gene 2027 |
759 |
0.45 |
| chr12_52478996_52479167 | 0.15 |
Gm35135 |
predicted gene, 35135 |
1528 |
0.39 |
| chr14_11716901_11717053 | 0.15 |
Gm48602 |
predicted gene, 48602 |
50966 |
0.16 |
| chr11_50733265_50733435 | 0.14 |
Gm12200 |
predicted gene 12200 |
7806 |
0.16 |
| chr5_28465358_28465587 | 0.14 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
1512 |
0.34 |
| chr2_30378803_30378994 | 0.14 |
Miga2 |
mitoguardin 2 |
663 |
0.51 |
| chr11_88618341_88618520 | 0.14 |
Msi2 |
musashi RNA-binding protein 2 |
28283 |
0.2 |
| chr10_93196295_93196461 | 0.14 |
Cdk17 |
cyclin-dependent kinase 17 |
10609 |
0.2 |
| chrX_60322703_60322854 | 0.14 |
Atp11c |
ATPase, class VI, type 11C |
55948 |
0.12 |
| chr13_4433467_4433618 | 0.14 |
Akr1c6 |
aldo-keto reductase family 1, member C6 |
764 |
0.63 |
| chr13_56052991_56053142 | 0.14 |
Gm47072 |
predicted gene, 47072 |
22832 |
0.16 |
| chr14_21114751_21114902 | 0.14 |
Adk |
adenosine kinase |
38674 |
0.17 |
| chr9_122056710_122056883 | 0.13 |
Gm39465 |
predicted gene, 39465 |
5333 |
0.13 |
| chr12_101953826_101954166 | 0.13 |
Atxn3 |
ataxin 3 |
4152 |
0.17 |
| chr7_87246956_87247377 | 0.13 |
Nox4 |
NADPH oxidase 4 |
354 |
0.89 |
| chr6_54587839_54588008 | 0.13 |
Fkbp14 |
FK506 binding protein 14 |
1822 |
0.3 |
| chr18_70573928_70574153 | 0.13 |
Mbd2 |
methyl-CpG binding domain protein 2 |
5706 |
0.21 |
| chr4_100397682_100397872 | 0.13 |
Gm12706 |
predicted gene 12706 |
35718 |
0.19 |
| chr13_47237173_47237324 | 0.13 |
Rnf144b |
ring finger protein 144B |
43423 |
0.14 |
| chr5_89243570_89243737 | 0.13 |
Gc |
vitamin D binding protein |
191975 |
0.03 |
| chr14_55459733_55459946 | 0.13 |
Dhrs4 |
dehydrogenase/reductase (SDR family) member 4 |
18919 |
0.09 |
| chr4_97820602_97820781 | 0.12 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
42613 |
0.16 |
| chr17_53588113_53588335 | 0.12 |
Kat2b |
K(lysine) acetyltransferase 2B |
4098 |
0.18 |
| chr8_113140329_113140534 | 0.12 |
Gm10280 |
predicted gene 10280 |
73169 |
0.13 |
| chr2_34049474_34049625 | 0.12 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr7_79740953_79741105 | 0.12 |
Gm44706 |
predicted gene 44706 |
351 |
0.6 |
| chr6_117297591_117297939 | 0.12 |
Rpl28-ps4 |
ribosomal protein L28, pseudogene 4 |
83699 |
0.08 |
| chr4_137351449_137351600 | 0.12 |
Cdc42 |
cell division cycle 42 |
6175 |
0.14 |
| chr15_36832238_36832450 | 0.12 |
Gm49282 |
predicted gene, 49282 |
10472 |
0.16 |
| chr15_102752976_102753138 | 0.12 |
Calcoco1 |
calcium binding and coiled coil domain 1 |
30879 |
0.11 |
| chr7_107699185_107699336 | 0.12 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
33949 |
0.12 |
| chr19_21487417_21487631 | 0.12 |
Gda |
guanine deaminase |
14079 |
0.24 |
| chr6_79885576_79885747 | 0.12 |
Gm20594 |
predicted gene, 20594 |
67745 |
0.12 |
| chr10_85161219_85161370 | 0.12 |
Cry1 |
cryptochrome 1 (photolyase-like) |
9938 |
0.18 |
| chr4_152741969_152742142 | 0.12 |
Gm833 |
predicted gene 833 |
44515 |
0.17 |
| chr2_78656286_78656827 | 0.12 |
Gm14463 |
predicted gene 14463 |
873 |
0.71 |
| chr4_88889115_88889274 | 0.12 |
Ifne |
interferon epsilon |
8993 |
0.08 |
| chr16_58629623_58629774 | 0.12 |
Ftdc2 |
ferritin domain containing 2 |
9041 |
0.14 |
| chr13_95326893_95327258 | 0.12 |
Zbed3 |
zinc finger, BED type containing 3 |
308 |
0.86 |
| chr9_95562380_95562552 | 0.11 |
Paqr9 |
progestin and adipoQ receptor family member IX |
2809 |
0.19 |
| chr2_165869197_165869348 | 0.11 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
6505 |
0.15 |
| chr5_137986079_137986259 | 0.11 |
Azgp1 |
alpha-2-glycoprotein 1, zinc |
4601 |
0.1 |
| chr1_188652846_188653036 | 0.11 |
Gm25095 |
predicted gene, 25095 |
29888 |
0.23 |
| chr8_85595702_85595890 | 0.11 |
Gm45424 |
predicted gene 45424 |
4036 |
0.19 |
| chr9_79786091_79786242 | 0.11 |
Tmem30a |
transmembrane protein 30A |
7264 |
0.15 |
| chr7_133463081_133463245 | 0.11 |
Gm45672 |
predicted gene 45672 |
129962 |
0.04 |
| chr2_119586949_119587118 | 0.11 |
Oip5os1 |
Opa interacting protein 5, opposite strand 1 |
7264 |
0.12 |
| chr4_58508983_58509134 | 0.11 |
Lpar1 |
lysophosphatidic acid receptor 1 |
9655 |
0.2 |
| chr11_4833141_4833320 | 0.11 |
Nf2 |
neurofibromin 2 |
97 |
0.96 |
| chr6_117905744_117905933 | 0.11 |
Hnrnpf |
heterogeneous nuclear ribonucleoprotein F |
952 |
0.37 |
| chr2_103626307_103626554 | 0.11 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
60120 |
0.1 |
| chr19_33098550_33098701 | 0.11 |
Gm29946 |
predicted gene, 29946 |
24022 |
0.18 |
| chr6_47466848_47467049 | 0.10 |
Cul1 |
cullin 1 |
12560 |
0.17 |
| chr15_97228689_97229019 | 0.10 |
Pced1b |
PC-esterase domain containing 1B |
18253 |
0.19 |
| chr7_98308016_98308180 | 0.10 |
Acer3 |
alkaline ceramidase 3 |
1408 |
0.44 |
| chr1_58203680_58204225 | 0.10 |
Aox4 |
aldehyde oxidase 4 |
6445 |
0.19 |
| chr14_100225710_100225880 | 0.10 |
Gm16260 |
predicted gene 16260 |
6309 |
0.21 |
| chr13_51232215_51232383 | 0.10 |
Gm29787 |
predicted gene, 29787 |
26188 |
0.14 |
| chr2_17186295_17186446 | 0.10 |
Gm13322 |
predicted gene 13322 |
883 |
0.67 |
| chr4_49459901_49460052 | 0.10 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
8839 |
0.13 |
| chr17_70845881_70846072 | 0.10 |
Tgif1 |
TGFB-induced factor homeobox 1 |
350 |
0.78 |
| chr7_30191689_30191840 | 0.10 |
Capns1 |
calpain, small subunit 1 |
646 |
0.42 |
| chr10_61666021_61666218 | 0.10 |
Ppa1 |
pyrophosphatase (inorganic) 1 |
2430 |
0.19 |
| chr12_116772730_116773138 | 0.10 |
Gm48119 |
predicted gene, 48119 |
80168 |
0.1 |
| chr8_69954561_69954952 | 0.10 |
Gatad2a |
GATA zinc finger domain containing 2A |
3648 |
0.16 |
| chr16_66005549_66005730 | 0.10 |
Gm49635 |
predicted gene, 49635 |
58533 |
0.15 |
| chr9_92265880_92266265 | 0.10 |
Plscr1 |
phospholipid scramblase 1 |
223 |
0.88 |
| chr5_150228070_150228236 | 0.10 |
Gm36378 |
predicted gene, 36378 |
15710 |
0.19 |
| chr17_93509221_93509442 | 0.10 |
Gm50001 |
predicted gene, 50001 |
117633 |
0.06 |
| chr8_93248271_93248422 | 0.10 |
Gm45727 |
predicted gene 45727 |
2829 |
0.19 |
| chr1_61649748_61649945 | 0.10 |
Gm37205 |
predicted gene, 37205 |
4577 |
0.17 |
| chr2_135853170_135853365 | 0.10 |
Plcb4 |
phospholipase C, beta 4 |
34427 |
0.18 |
| chr10_63245966_63246166 | 0.10 |
Herc4 |
hect domain and RLD 4 |
244 |
0.86 |
| chr7_112224273_112224445 | 0.10 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
1497 |
0.52 |
| chr12_104034678_104035206 | 0.10 |
Serpina12 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
9501 |
0.1 |
| chr7_65367609_65368266 | 0.10 |
Gm44794 |
predicted gene 44794 |
1155 |
0.47 |
| chr18_50133917_50134085 | 0.10 |
Cd63-ps |
CD63 antigen, pseudogene |
725 |
0.66 |
| chr6_88046142_88046293 | 0.10 |
Rab7 |
RAB7, member RAS oncogene family |
947 |
0.41 |
| chr3_138336264_138336545 | 0.10 |
Adh6b |
alcohol dehydrogenase 6B (class V) |
5371 |
0.13 |
| chr14_60684364_60684529 | 0.10 |
Spata13 |
spermatogenesis associated 13 |
7529 |
0.24 |
| chr14_60387250_60387782 | 0.09 |
Amer2 |
APC membrane recruitment 2 |
9230 |
0.22 |
| chr10_68327935_68328096 | 0.09 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
6873 |
0.22 |
| chr17_55903412_55903563 | 0.09 |
Zfp959 |
zinc finger protein 959 |
11019 |
0.1 |
| chr3_49778482_49778633 | 0.09 |
Gm37550 |
predicted gene, 37550 |
2330 |
0.33 |
| chr3_18463182_18463725 | 0.09 |
Gm30667 |
predicted gene, 30667 |
2199 |
0.33 |
| chr5_121431395_121431558 | 0.09 |
Naa25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
7942 |
0.1 |
| chrX_113007609_113007802 | 0.09 |
Gm14936 |
predicted gene 14936 |
2049 |
0.28 |
| chr3_86821532_86821703 | 0.09 |
Dclk2 |
doublecortin-like kinase 2 |
22457 |
0.17 |
| chr19_20614762_20614977 | 0.09 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
12908 |
0.21 |
| chr5_147748468_147748664 | 0.09 |
Gm43156 |
predicted gene 43156 |
3050 |
0.26 |
| chr4_81505880_81506190 | 0.09 |
Gm11765 |
predicted gene 11765 |
44303 |
0.17 |
| chr1_24005783_24005985 | 0.09 |
Sdhaf4 |
succinate dehydrogenase complex assembly factor 4 |
228 |
0.66 |
| chr4_86897861_86898116 | 0.09 |
Acer2 |
alkaline ceramidase 2 |
23574 |
0.19 |
| chr5_117096808_117097327 | 0.09 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
1744 |
0.29 |
| chr17_32493234_32493405 | 0.09 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
1161 |
0.38 |
| chr19_41418415_41418566 | 0.09 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
33388 |
0.16 |
| chr2_109757612_109757763 | 0.09 |
Gm13932 |
predicted gene 13932 |
34847 |
0.16 |
| chr6_23645311_23645468 | 0.09 |
Rnf133 |
ring finger protein 133 |
4817 |
0.19 |
| chr6_15811355_15811665 | 0.09 |
Gm43924 |
predicted gene, 43924 |
187 |
0.96 |
| chr19_28834565_28834716 | 0.09 |
Slc1a1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
409 |
0.82 |
| chr13_101016552_101016703 | 0.09 |
Gm6114 |
predicted gene 6114 |
2641 |
0.28 |
| chr6_143133276_143133427 | 0.09 |
Gm44306 |
predicted gene, 44306 |
1736 |
0.28 |
| chr5_113217277_113217438 | 0.09 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
3879 |
0.17 |
| chr6_8862762_8862913 | 0.09 |
Ica1 |
islet cell autoantigen 1 |
84349 |
0.1 |
| chr14_27336610_27336992 | 0.09 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
735 |
0.71 |
| chr13_46466566_46466717 | 0.09 |
Cap2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
35207 |
0.14 |
| chr6_8350600_8350751 | 0.09 |
Gm16055 |
predicted gene 16055 |
13461 |
0.18 |
| chr15_52161435_52161609 | 0.09 |
Gm2387 |
predicted gene 2387 |
72214 |
0.1 |
| chr2_156393156_156393343 | 0.09 |
2900097C17Rik |
RIKEN cDNA 2900097C17 gene |
270 |
0.81 |
| chr8_71699244_71699423 | 0.09 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
2456 |
0.13 |
| chr8_106599394_106599703 | 0.09 |
Cdh1 |
cadherin 1 |
3803 |
0.2 |
| chr1_70927826_70928016 | 0.09 |
Gm16236 |
predicted gene 16236 |
112109 |
0.07 |
| chr19_37177758_37177920 | 0.08 |
Cpeb3 |
cytoplasmic polyadenylation element binding protein 3 |
178 |
0.9 |
| chr18_64598484_64598811 | 0.08 |
Gm50338 |
predicted gene, 50338 |
2996 |
0.22 |
| chr9_70062078_70062269 | 0.08 |
Gm47233 |
predicted gene, 47233 |
7694 |
0.14 |
| chr12_79192021_79192196 | 0.08 |
Rdh11 |
retinol dehydrogenase 11 |
185 |
0.9 |
| chr16_31405158_31405321 | 0.08 |
Gm49736 |
predicted gene, 49736 |
7228 |
0.13 |
| chr3_41021798_41021949 | 0.08 |
Larp1b |
La ribonucleoprotein domain family, member 1B |
66 |
0.96 |
| chr9_20467322_20467525 | 0.08 |
Zfp426 |
zinc finger protein 426 |
6039 |
0.11 |
| chr9_43825824_43825990 | 0.08 |
Nectin1 |
nectin cell adhesion molecule 1 |
3924 |
0.23 |
| chr4_23729753_23729929 | 0.08 |
Gm11890 |
predicted gene 11890 |
69497 |
0.13 |
| chr2_75999553_75999704 | 0.08 |
Pde11a |
phosphodiesterase 11A |
6415 |
0.18 |
| chr6_143597549_143597700 | 0.08 |
1700060C16Rik |
RIKEN cDNA 1700060C16 gene |
53664 |
0.15 |
| chr9_104263375_104263551 | 0.08 |
Dnajc13 |
DnaJ heat shock protein family (Hsp40) member C13 |
533 |
0.67 |
| chr11_62487235_62487386 | 0.08 |
Gm12278 |
predicted gene 12278 |
4513 |
0.13 |
| chr5_25222534_25222916 | 0.08 |
Galnt11 |
polypeptide N-acetylgalactosaminyltransferase 11 |
157 |
0.78 |
| chr11_86681158_86681454 | 0.08 |
Vmp1 |
vacuole membrane protein 1 |
2515 |
0.22 |
| chr8_116364517_116364668 | 0.08 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
7531 |
0.3 |
| chr15_80876034_80876209 | 0.08 |
Tnrc6b |
trinucleotide repeat containing 6b |
4577 |
0.22 |
| chr6_72627194_72627563 | 0.08 |
Gm15401 |
predicted gene 15401 |
1910 |
0.15 |
| chr11_4571360_4571745 | 0.08 |
Gm11960 |
predicted gene 11960 |
7920 |
0.16 |
| chr9_44940562_44940729 | 0.08 |
Gm19121 |
predicted gene, 19121 |
15070 |
0.08 |
| chr15_75310862_75311190 | 0.08 |
Gm3454 |
predicted gene 3454 |
3938 |
0.12 |
| chr1_79822993_79823175 | 0.08 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
1762 |
0.36 |
| chr7_29859607_29859795 | 0.08 |
Zfp420 |
zinc finger protein 420 |
278 |
0.79 |
| chr3_133750379_133750596 | 0.08 |
Gm6135 |
prediticted gene 6135 |
41017 |
0.15 |
| chr12_40032556_40032707 | 0.08 |
Arl4a |
ADP-ribosylation factor-like 4A |
4736 |
0.21 |
| chr2_126950478_126950775 | 0.08 |
Sppl2a |
signal peptide peptidase like 2A |
17391 |
0.16 |
| chr5_123709706_123709870 | 0.08 |
Zcchc8 |
zinc finger, CCHC domain containing 8 |
1306 |
0.31 |
| chr5_129930268_129930468 | 0.08 |
Vkorc1l1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
11602 |
0.09 |
| chr13_110421353_110421528 | 0.08 |
Plk2 |
polo like kinase 2 |
23643 |
0.22 |
| chr14_56604710_56604899 | 0.08 |
Gm16573 |
predicted gene 16573 |
15693 |
0.14 |
| chr6_136522266_136522417 | 0.08 |
Gm44140 |
predicted gene, 44140 |
3310 |
0.17 |
| chr2_130894667_130894834 | 0.08 |
A730017L22Rik |
RIKEN cDNA A730017L22 gene |
11606 |
0.13 |
| chr10_53319928_53320079 | 0.08 |
Pln |
phospholamban |
17664 |
0.12 |
| chr14_21411001_21411208 | 0.08 |
Gm25864 |
predicted gene, 25864 |
39370 |
0.14 |
| chr19_59642704_59642875 | 0.08 |
Gm19956 |
predicted gene, 19956 |
18534 |
0.21 |
| chr15_23106610_23106793 | 0.08 |
Gm24656 |
predicted gene, 24656 |
48033 |
0.17 |
| chr2_75883655_75883819 | 0.08 |
Agps |
alkylglycerone phosphate synthase |
9781 |
0.18 |
| chr4_32656586_32656987 | 0.08 |
Mdn1 |
midasin AAA ATPase 1 |
333 |
0.7 |
| chr12_108235642_108235812 | 0.08 |
Ccnk |
cyclin K |
36642 |
0.13 |
| chr7_79907099_79907250 | 0.08 |
Gm26646 |
predicted gene, 26646 |
9763 |
0.1 |
| chrX_164445705_164445870 | 0.08 |
Asb11 |
ankyrin repeat and SOCS box-containing 11 |
7729 |
0.17 |
| chr11_81370078_81370386 | 0.08 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
14781 |
0.29 |
| chr2_69199630_69199898 | 0.08 |
Spc25 |
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
5366 |
0.17 |
| chr17_80786074_80786840 | 0.08 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
57972 |
0.12 |
| chr9_78007022_78007181 | 0.08 |
Gm23609 |
predicted gene, 23609 |
6997 |
0.12 |
| chr7_70350632_70350952 | 0.07 |
Gm44948 |
predicted gene 44948 |
3096 |
0.16 |
| chr1_170485088_170485365 | 0.07 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
30312 |
0.19 |
| chr9_65330615_65330808 | 0.07 |
Gm39363 |
predicted gene, 39363 |
1809 |
0.18 |
| chr4_129888230_129888411 | 0.07 |
Gm23845 |
predicted gene, 23845 |
7253 |
0.12 |
| chr3_10466008_10466168 | 0.07 |
Snx16 |
sorting nexin 16 |
25986 |
0.18 |
| chr14_75798160_75798317 | 0.07 |
Slc25a30 |
solute carrier family 25, member 30 |
11201 |
0.14 |
| chr12_35049764_35050116 | 0.07 |
Snx13 |
sorting nexin 13 |
2723 |
0.27 |
| chr10_45070700_45070964 | 0.07 |
Prep |
prolyl endopeptidase |
3629 |
0.22 |
| chr9_71768236_71768387 | 0.07 |
Cgnl1 |
cingulin-like 1 |
3249 |
0.25 |
| chr10_89563955_89564134 | 0.07 |
Gm48087 |
predicted gene, 48087 |
8325 |
0.2 |
| chr9_7987133_7987475 | 0.07 |
Yap1 |
yes-associated protein 1 |
14399 |
0.15 |
| chr2_64072797_64073328 | 0.07 |
Fign |
fidgetin |
24926 |
0.28 |
| chr1_62423315_62423491 | 0.07 |
Pard3bos3 |
par-3 family cell polarity regulator beta, opposite strand 3 |
75858 |
0.1 |
| chr8_124499720_124499883 | 0.07 |
Cog2 |
component of oligomeric golgi complex 2 |
20966 |
0.17 |
| chr16_76432143_76432294 | 0.07 |
Gm9843 |
predicted gene 9843 |
28566 |
0.17 |
| chr10_108509674_108510023 | 0.07 |
Gm47466 |
predicted gene, 47466 |
27198 |
0.2 |
| chr5_52993515_52993725 | 0.07 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
816 |
0.57 |
| chr7_62923766_62924082 | 0.07 |
A26c2 |
ANKRD26-like family C, member 2 |
100251 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0052428 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0018656 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |